服务
文章信息
- 刘永平, 杨静, 杨明峰
- Yongping Liu, Jing Yang, and Mingfeng Yang
- 植物开花调控途径
- Pathways of flowering regulation in plants
- 生物工程学报, 2015, 31(11): 1553-1566
- Chin J Biotech, 2015, 31(11): 1553-1566
- 10.13345/j.cjb.140626
-
文章历史
- Received:December 18, 2014
- Accepted:February 5, 2015
;2 北京农学院 农业部都市农业 (北方) 重点开放实验室,北京 102206
2. Key Laboratory of Urban Agriculture (North) of Ministry of Agriculture China, Beijing University of Agriculture, Beijing 102206,China
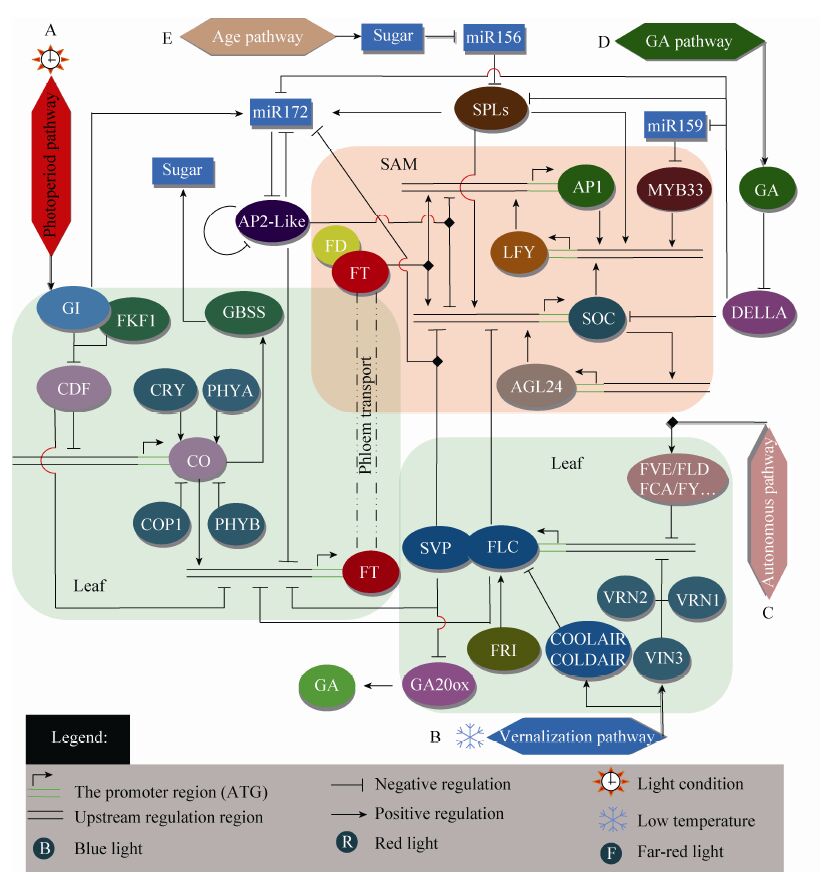
高等植物花发育经历开花诱导 (The floral induction phase)、花原基形成 (The floral primodia phase) 和花器官发育 (The floral organs development phase) 3个阶段,相对应的调控基因包括:开花时间基因 (Flowering time genes)、 分生组织特征基因 (Metristem identity genes) 和花器官特征基因 (Organ identity genes)。而其中的开花诱导阶段,虽然植物的茎顶端分生组织 (Shoot apical meristem,SAM) 在形态上没有发生变化,但是以FT (Flowering locus T) 和SOC1 (Suppressor of overexpression of CO 1) 为中心的系列开花基因正在发生着各种有规律的变化,形成一个复杂的调控网络,以实现植物从营养生长向生殖生长的不可逆转变,是植物个体发育和后代繁衍的关键阶段。对开花诱导调控网络的研究,将响应外界环境刺激和内源信号启动开花途径分为:光周期途径 (Photoperiod pathway)、春化途径 (Vernalization pathway)、自发途径 (Autonomous pathway)、赤霉素途径 (Gibberellin pathway,GA pathway)、和年龄途径 (Age pathway)。
1 光周期途径 1.1 长日植物CO/FT模式光周期是一种约24 h生物时钟节律 (Circandian rhythm,circadian clock),即植物根据光照时间的周期性变化来协调自身新陈代谢及各种生理过程,最终实现开花诱导、叶片运动和气孔开闭等生长发育变化。以拟南芥 Arabidopsi thaliana 为代表的长日植物研究显示:CO (Constans)/FT模式的表达变化是光周期诱导途径中最核心的环节[1]。
CO是典型的时钟控制基因,编码一个包含2 个锌指结构 (B-box) 的光稳定蛋白,依赖N端区域调节蛋白与蛋白之间的互作,在C末端有核定位所需要的CCT (CO,CO-like,timing of CAB expression 1) 区域,属于一类没有明显DNA 结合区域的转录调控因子,作用于FT 启动子区域,是FT主要的正调控因子[2]。叶片中FT 编码的成花因子FT (Florigena) 通过韧皮部从叶片到达SAM,与bZIP锌指蛋白FD结合上调下游的花分生组织特征基因AP1 (Apetala1) 和SOC1的表达,再由它们调控其他的花器官特征基因LFY (Leafy) 和AG24 (Agamous-Like 24) 等,最终完成花器官发育[1, 2]。
CDFs (Cycling DOF factor) 是一类植物特有的单锌指DOF转录因子,表达也受时钟控 制[3, 4]。CDF1能结合到CO启动子区抑制CO表达,过量表达AtCDF1 的拟南芥晚花,表明其是开花的负调控因子。它被F-box蛋白 (Flavin- binding kelch-repeat 1,FKF1) 和GI (Gigantea) 形成的FKF1-GI复合体标记后降解。简而言之,在长日照条件下,FKF1和GI的表达正午达到同步高峰,随后形成稳定的FKF1-GI蛋白复合体,与位于CO启动子区的CDF1结合,随后被泛素化蛋白酶降解,解除对CO的抑制作用,保证CO午后启动FT的表达,最终诱导开 花[4, 5]。当然,CDF1除了调控CO外,也可以直接结合到FT的启动子区负调控FT的表达,抑制开花[6] (图1A)。另外,一种植物特有的Embryonic Flower1 (EMF1)、Like herochromain protein1 (LHP1) 和一个组蛋白H3K4去甲基酶形成一个多梳复合体 (Polycomb group,PcG) 即EMF1c,该复合体只在长日条件下的午后受到CO的影响,从FT启动子远端解离,调控FT在黄昏前达到高峰,促进拟南芥在长日照下 开花[7]。
|
| 图1 开花调控途径:光周期途径 (A)、春化途径 (B)、自主开花途径 (C)、赤霉素途径 (D) 和年龄途径 (E) Fig.1 The main flowering pathways. (A) Photoperiod pathway. (B) Vernalization pathway. (C) Autonomous pathway. (D) Gibberellins pathway. (E) Age pathway. |
CO除了调控FT外,还通过调控淀粉粒结合型淀粉合成酶GBSS (Granule-bound starch synthetase) 的表达水平和表达时间,影响光周期响应的淀粉平衡,这对于开花转换需要的可溶性糖增加非常关键[8]。另一个糖代谢基因——蔗糖合成酶基因SUS4 (Sucrose synthase) 受到光周期和糖依赖基AtIDD8 (Indeterminate domain) 的双重影响,增加的蔗糖继续促进AtIDD8-SUS4通路,最终实现代谢稳态和光周期开花的平衡[9]。由此也说明光周期开花与糖代谢关系密切。
不仅光周期,光质也能影响CO/FT模式的稳定性。GI受蓝光调控,还可以分别与另外两个泛素蛋白ZTL (Zeitlupe) 和LKP2 (Lov kelch repeat protein 2) 结合,降解其他的AtCDF2,3,5,共同调控拟南芥的开花[10, 11]。CO蛋白稳定性既受到蓝光受体隐花色素CRY1、CRY2 (Cryptochromes) 和红光/远红光受体光敏色素PHYA (Phytochromes A) 的正调控[2],又受到PHYB[2]、E3泛素化蛋白COP1 (Constitutive photomorphogenesis 1)[12]和FKF1的负调控[6] (图1A)。
1.2 CO/FT模式的保守性短日植物水稻Oryza sativa的HD1 (Heading date 1) 是CO同源基因,在短日照条件下HD1促进FT同源基因Hd3a (Heading date 3a) 表达,呈现早花[13],但在长日照条件下却抑制Hd3a。Lifschitz团队证明日中性植物西红柿Solanum lycopersicum的CO同源基因TCOL1,3 (Tomato constans like) 属于时钟控制基因,响应光周期呈现节律性表达变化,但是与开花时间无关[14];FT同源基因SFT (Single flower truss) 不仅启动西红柿开花,还诱导生长的停止;但是这种控制与日照长短无关[15]。在裸子植物云杉 Picea asperata [16]、多年生杨树Populus tremula [17]、洋葱Allium cepa[18]和马铃薯Solanum tuberosum [19]中也发现FT同源基因的表达不仅诱导开花,还与启动休眠、鳞芽形成和结薯密切关联。
水稻OsRdd1 (AtCDF的同源基因) 受到光质和时钟节律的控制,过量表达反义OsRdd1-AS的水稻表现晚花、植株矮小和种子变小[20]。Li等[21]也指出过量表达OsDof12的水稻 (在Iwamoto等[20]中对应的是OsRdd4) 在长日照条件下早花,说明短日植物OsRdd1和OsDof12 也表现出类似的调控开花时间的作用。我们从日中性植物麻疯树中克隆到JcDof1、JcDof3和另外3个由数据库http://www.kazusa.or.jp/jatropha/筛选到的JcDof4-6基因 (资料略) 表达呈现有规律的时钟振颤,其中JcDof1、JcDof3能与GI在酵母中发生互作,但是不清楚它们是否对开花时间有影响[22, 23, 24]。
上述研究显示不同植物中的CO/FT模式存在一定保守性,但是在组件构成及调控通路上还存在差异。特别是对于光周期不敏感植物仍保留相关同源基因,但却表现出与光周期无关的开花现象仍需深入研究。
2 春化途径春化作用 (Vernalization) 是植物适应季节变化,在一定低温 (一般为4 ℃下处理2−8周) 处理后,表现出对越冬植物成花的诱导和促进作用。如果这些植物不经低温处理,开花过程可能推迟几周甚至几个月。春化作用的具体效应取决于植物所处的发育阶段、低温处理时间长短和温度等几个因素[25]。
2.1 双子叶植物的春化途径目前,双子叶植物拟南芥春化途径的分子机制研究主要集中在FLC基因的表达变化。FLC (Flower locus C) 是MADS-box类蛋白,它在叶中抑制FT基因的表达,在SAM中抑制FD和SOC1的表达,是开花的强抑制因子[26]。FRI (Frigida) 是FLC的正调控因子,不受低温诱导,促进FLC高水平表达进而抑制开花,FRI基因突变能引起早花[27]。VRN1 (Vernalization 1)、VRN2 (Vernalization 2) 和VIN3 (Vernalization insensitive3) 这3个基因的产物属于FLC的负调控因子。VRN1编码一种植物特有的非特异性DNA结合蛋白,参与染色质结构的改变[28]。VRN2是第一个被克隆的春化作用基因,编码一个核定位的锌指蛋白,功能类似于多梳蛋白 (PcG)[29]。它们在不同组织和不同发育阶段均有表达,不受低温诱导,作用是抑制FLC在春化作用解除后的重复增加,保证春化作用的稳定性,促进开花[28, 29, 30]。VIN3编码一类PHD (Plant homeodomian) 锌指结构蛋白,受春化作用诱导,参与FLC基因染色质重塑 (Chromatin remodeling) 即组蛋白H3K9和H3K27甲 基化,使其从“激活”转为“抑制”状态。它的诱导性保证了FLC的表达在后代胚胎发育阶段被重置,以确保春化作用的重新获得[30, 31]。前面提到的多梳蛋白复合体EMF1c中的EMF1被证明能在酵母中与FLC和FLM (Flowering locus M,a FLC-clade member) 相互作用,推测EMF1-FLC-FLM可能形成多重复合体抑制FT 表达[7, 32]。
与长链非编码RNA (Long non-coding RNA,lncRNA) 有关的春化作用研究发现:低温诱导的长链反义RNA (COOLAIR,Cool induced long antisense RNA) 和冷辅助内含子非编码RNA (COLDAIR,Cold assisted intronic noncoding RNA) 引起FLC转录水平下调促进开花。第一种COOLAIR,由RNA聚合酶Ⅱ (RNA polymeraseⅡ,PolⅡ) 转录,是植物体内天然存在的FLC反向转录本,具有典型的5′帽子结构和3′端聚腺苷酸 (poly A) 结构[33]。在春化过程中,FLC编码区3′端下游启动子启动COOLAIR表达。根据剪切方式的不同,COOLAIR分为近3′ 端和远3′端两种类型,都具有两个多聚腺苷酸化位点。远端COOLAIR的聚腺苷酸化维持FLC的高表达;而近端与FLC的低表达相关[33, 34]。如果低温诱导的近端COOLAIR表达升高受到干扰,增加的FLC又会通过反馈作用间接增加COOLAIR,重新抑制FLC,最终调控FLC的量[35]。第二种 COLDAIR,由FLC正义链第1个内含子形成,其转录本长1.1 kb,具有5′帽子结构,但是缺少3′ poly A结构。最新发现,与COLDAIR有利于多梳蛋白抑制复合体2 (Plant homeodomain polycomb repressive complex2,PHD-PRC2) 和FLC组蛋白结合促进H3K27me3甲基化不同[36],COOLAIR是通过促进组蛋白不同位点H3K36me3甲基化实现对FLC的抑制,它们是两条相对独立的途径[37]。总之,随着低温处理时间的加长,COOLAIR、COLDIR和VIN3的表达水平依次达到高峰,而开花抑制因子FLC则逐渐减少[36] (图1B)。
随着春化作用的研究不断深入,FLC的同源基因在其他作物中相继被克隆。大白菜Brassica chinensis的BrFLC1、BrFLC2和BrFLC3和甜菜 Beta vulgaris中BvFL1都是FLC-like的同源基因,把它们分别转化到拟南芥flc突变体后,恢复了开花抑制[38, 39]。但是菊苣 Cichorium intybus中的FLC-like基因CiMFL,未恢复flc突变体的表型[40],说明在十字花科植物中FLC功能的保守程度及其在春化途径中的作用仍存在差异。
2.2 单子叶植物的春化途径单子叶植物小麦 Triticum aestivum的研究发现,小麦中的3个春化作用必需基因是VRN1、VRN2和VRN3,都不是拟南芥VRN的同源物,也不包含FLC-like基因,小麦可能采取不同于拟南芥依赖的FLC的春化途径[41, 42]。
小麦VRN1属于MADS-box转录因子,与拟南芥花器官基因AP1、FUL (Fruitfull) 和CAL (Cauliflower) 同源,受春化作用诱导,是开花的必要条件[41]。VRN2属于CCT (Constans,constans-like,timing of CAB expression 1) 蛋白,在拟南芥中无同源物,是一个长日 (未春化) 条件下开花的负调控因子,阻碍小麦在夏秋开花[43]。短日植物水稻中VRN2的同源物是Ghd7,也显示长日照下抑制开花[44]。Yan等[45]发现大麦 Hordeum vulgare 和小麦中的春化基因VRN3是FT基因的同源物,在春化过程中作用类似拟南芥VIN3起传递春化信号作用。综合来看:长日 (未春化) 条件下VRN2大量表达抑制了VRN3,但是经历春化作用后,这种抑制随着VRN1的诱导表达被消除,增加的VRN3通过与FDL2 (Flowering locus-like 2) 蛋白的相互作用刺激VRN1的大量表达,实现开花诱导[41, 42, 43, 45] (图2)。它们3个的反馈环路,整合了春化作用和光周期信号诱导开花,说明尽管拟南芥与单子叶植物的春化途径中存在一些蛋白结构域及表观调控的相似之处,但在这两类植物中春化途径的组件构成和响应机制上差异较大,可能是独立进化的。
3 自主开花途径外界环境因素对植物开花的诱导可使植物在比较适宜的环境下开花,但如果缺少光信号、温度信号的诱导,植物通过感受自身内部的发育状态,在营养生长到达一定阶段后也会开花。迄今为止发现的拟南芥开花突变体中还没有完全不开花的,这说明植物内部还存在着控制开花的自主途径。比较清楚的自主途径也是通过抑制FLC基因的表达来促进开花的。在拟南芥中已经相继克隆到FCA (Flower locus CA)、FY (Flower locus Y)、FPA (Flower locus PA)、FVE (Flower locus VE)、FLD (Flower locus D)、FLK (Flower locus K Homology domain) 和LD (Luminidependens) 等7个基因,它们以不同方式参与对FLC染色质或其mRNA的修饰与调 节[25, 46]。FCA、FPA、FLK和FY基因都编码RNA结合蛋白,它们参与FLC前体mRNA的调节,在开花控制中非常关键,属于转录后调控[25, 34]。譬如FCA、FY和FPA会促进COOLAIR转录本近3′端多聚腺苷酸化位点的形成,引起FLC的沉默,有利于开花[34, 47],但是FCA和FPA对3′端多聚腺苷酸位点的选择采取相互独立的模式,呈现出对FLC的相对冗余的抑制作用[47]。LD、FLD和FVE编码调节FLC表观遗传因子,参与FLC染色质组蛋白去乙酰化的后期修饰,其中FVE还受低温诱导[25, 48, 49],这说明自主途径与春化途径共同通过调整染色质结构 (如甲基化和去乙酰化等) 来控制FLC的表达 (图1C)。
4 赤霉素途径赤霉素 (Gibberellins,GAs) 是一种二萜酸类植物激素,在包括种子萌发、茎杆伸长、花粉成熟、开花诱导和花器官形成等方面发挥重要作用。目前从植物、真菌和细菌中鉴定出来的GAs超过130种,但大多数都是作为有生物活性形式的前体 (无生物活性形式) 存在,其中有活性的GAs主要有GA1、GA3、GA4和 GA7[50]。
4.1 GA调控开花在非诱导的短日照条件下,GA是拟南芥开花的必要条件,而GA处理可以促进短日照下拟南芥早花[51]。2005年Ueguchi-Tanaka等[52]在水稻中分离获得赤霉素受体GID1 (Gibberellin insensitive dwarf 1),之后在拟南芥中也获得了3个同源的GID1基因AtGID1a、AtGID1b和AtGID1c。GA和GID1结合后可与一类C端非常保守、N端具有DELLA (Asp-Glu- Leu-Leu-Ala) 结构域的DELLA蛋白形成GA-GID1-DELLA三聚体[53]。拟南芥基因组中,目前共发现5个DELLA蛋白:RGA (Repressor of GA)、GAI (GA insensitive)、RGL1、RGL2和RGL3 (RGA-like),它们在序列和功能上都极为相似[53, 54]。研究表明上述DELLA基因如果发生缺失或突变,拟南芥均表现早花;而当GA合成途径被打断时,则表现为晚花。深入研究还发现GA-GID1-DELLA三聚体形成后被SCF (SKP1-CUL1-F-box) 聚合体标记,之后DELLA被泛素26S蛋白酶体降解,从而解除了DELLA蛋白对植物生长的抑制作用,产生赤霉素效 应[53, 54]。在GA调控拟南芥开花的途径中,主要是解除了DELLA对SPL9 (Squamosa promoter binding likes) 的抑制,有利于其下游LFY和SOC1的表达[55]。这样就明确了由泛素-蛋白酶体降解途径介导调节了GA转导的GA-GID1-DELLA信号通路,实现了对开花诱导的调节[54, 55] (图1D)。最新研究发现:GA既能终止营养生长,也会抑制花器官形成。GA促进LFY的表达,调控开花转换,但是增加的LFY却诱导了一个细胞色素P450的基因ELA1 (Eui-like P450) 在SAM中的表达,ELA1会阻碍GA碳骨架C13的羟基化,减少了GA4的合成,结果使依赖于GA-DELLA-AP1通路的花器官形成机制受到破坏,反而推迟花芽形成[56]。这说明GA在花芽形成上可能扮演既促进又抑制的双重角色,也为GA抑制木本植物花芽的形式,而GA抑制剂多效唑PCA (Paclobutrazol) 反而促进花芽形成和植株的矮化的现象[57, 58],提供了非常重要的研究思路。
ELF3 (Early flower 3) 是一个与光周期开花有关的时钟控制基因,近期关于大麦的研究中发现:在短日照下ELF3的表达量增加不仅抑制GA20ox (GA oxidase) 减少GA,还抑制FT1;但在长日照下,它的表达减少,解除对GA20ox和FT1的抑制,分别促进了花分生组织特征基因和MAD-box花器官特征基因表达[59],这说明GA 和光周期两条途径既独立又密切相关。
4.2 GA调控miRNA159miRNAs是一类长21−24 nt的非编码内源性单链RNA,广泛分布于动植物中。植物miRNA是由内切酶DCL1 (Dicer 1) 在核内切割后产生。成熟的miRNA运输到细胞质,然后包装进RNA诱导的沉默复合体RISC (RNA-induced silencing complex),通过转录切割、翻译抑制或DNA甲基化等方式调控基因表达[60]。miR159介导了与GA有关的开花调控,它通过抑制GA特异性转录因子MYB33、MYB65和MYB101 (R2R3-MYB转录因子) 发挥作用。在短日照条件下,GA不足,miR159增加,强烈抑制MYB33的表达;同时也抑制了MYB33下游基因LFY水平,导致拟南芥晚花。这种情况可以通过增加日长或者外源GA的喷施消除[55]。研究还发现GA-miR159- MYB33与GA-SOC1对开花的调控是彼此独立的两条途径[61]。有趣的是miR159a和miR159b及它们的靶基因MYB33和MYB65都存在既冗余又互补的现象[62]。其实,为了能保证植物在复杂环境中完成开花繁育的重任,植物开花调控中的很多环节,都扮演着既重要又可替代的角色。
5 年龄途径实际上,一年生植物在响应相应信号刺激转入生殖生长阶段之前也要经历一段时间的营养生长,只是比起要经历几年,甚至十几年幼年期的多年生植物而言要短得多。目前,关于从植物幼年期营养发育转入成年期生殖发育的研究主要集中在与miR156和miR172有关的级联式调控体系中[63, 64, 65]。
5.1 miR156-miR172的级联调控miR156和miR172几乎存在于所有植物类群,在拟南芥[64]、水稻[66]、弯曲碎米荠 Cardamine flexuosa[67]和杨树[68]等植物中均能找到。随着植物的生长发育,miR156逐渐减少,miR172逐渐增加,最终实现开花[64]。在转miR156的水稻[66]、番茄Solanum lycopersicum[69]和玉米Zea mays[70]等植物中,都能观察到晚花现象,而过量表达miR172则会导致严重的早花表型[64],表明miR172和miR156在植物开花调控中分别扮演了正、负调控因子的角色。
miR156的靶基因是一类称为SPLs的转录因子,含有SPB-box区,通过结合到开花器官特征基因启动子的SQUAMOSA区域进而促进开 花[63, 71]。在拟南芥基因组中,17个SPL基因中的11个是miR156的靶基因[63];而在水稻中,全部11个SPL均是miR156的靶基因[66];在小立碗藓中发现的SPB-box基因证实也是miR156的靶基因[71]。这些基因可分为两大类:SPL3和SPL9,SPL3比SPL9缺少C端的蛋白-蛋白相互作用 域[63, 64, 71]。在SAM中,SPL3和SPL9通过激活花器官特征基因AP1、LFY和SOC1直接诱导开花[63];在叶片中,SPL9通过激活miR172b,间接诱导开花[64]。miR172靶基因是AP2-like转录因子家族,包括AP2、SMZ (Schlafmutze)、SNZ (Schnarchzapfen)、TOE1、TOE2和 TOE3 (Target of eat),它们在被子植物、裸子植物及蕨类植物均有发现,都是FT基因的转录抑制子[64]。AP2-like一方面在叶片中直接负调控FT,一方面在SAM中抑制FT的下游基因SOC1和AP1;而且除了能反向负调控miRNA172和正调控miRNA156外,还能自我调节,在抑制开花中扮演比较活跃的角色[72]。系统的研究显示:miR156的表达水平逐渐下降的过程中SPLs水平逐渐升高,其中SPL9激活miR172,增加的miR172不断抑制靶基因AP2-like的表达,消除了它对FT、SOC1和AP1等基因的抑制,最终实现植物生长发育一段时间后转换为生殖生长[63, 72, 73],说明调控植物的开花过程中存在着精巧的miR156-miR172调控环 (图1E)。
5.2 调控miR172的其他途径miR172除了受到SPLs调控,还受到光周期途径中GI的调控。研究发现gi突变体中成熟miR172的水平明显减少,但初始miR172 (pri-miR172) 转录本不变,表明GI在miRNA的加工水平上促进miR172的积累,而且这条通路与GI-CO-FT途径是相对独立的[74]。自发途径中的关键基因FCA也能调控miR172,在23 ℃ (非正常温度16 ℃) 下,FCA的mRNA和蛋白水平都增高,它可能作为内切酶DCL1的一部分识别pri-miR172茎环结构的侧翼序列,加速剪切,从而增加成熟miR172的量,有利于拟南芥响应极端温度提早开花[75]。SVP (Short vegetative phase) 编码 MADS-box蛋白,主要功能是响应内源和外源信号抑制开花,它能与FLC互作分别负调控FT和SOC1[76];还能抑制GA20ox2,影响GA的合成,抑制开花[77]。最近发现SVP蛋白通过结合到miR172a启动子的CC(A/T)6GG(CArG) 区域上直接抑制miR172a的转录[78]。
5.3 糖信号与miR156为什么随着植物年龄的增长miR156会逐渐下降呢?目前,王佳伟[79]和Poethig[80]两个实验室分别通过“脱叶补糖法”证实:随着植物生长,光合碳代谢中糖的不断积累,特别是蔗糖和葡萄糖大大抑制了miR156a和miR156c的表达量,有利于植物从营养生长向生殖生长转换,并且糖对miR156的抑制在多种植物中表现出进化上保守性。结合前面讨论的响应光周期的几个糖代谢基因GBSS、AtIDD8-SUS4研究显示:糖是生殖转换的重要信号,处在植物营养水平和发育阶段的契合点上;也就是植物幼年期生长所积累的光合产物一部分作为糖、淀粉储藏起来,一部分作为信号传递出去,诱导生殖生长的开始。这暗示适合的植物C/N平衡对植物生殖转换的影响,值得深入研究。
这些与miRNA156和miR172有关的调控网络将相对独立的光周期、GA、自发途径和糖代谢交织到一起,共同实现从营养生长向生殖生长的精确转换。
6 小结与展望不断公布的新结果丰富了我们对植物如何从营养生长向生殖生长转换的理解,但是这些调控途径之间往往彼此独立又互相交织,形成一个复杂的网络体系,其中相对明确的观点包括:1) 开花本身不是植物从无到有的过程,而是其体内各类抑制因子逐渐解除的过程;譬如:长日照解除了CDF对CO和FT的抑制;春化作用和自发途径解除了FLC (或VRN3) 对FT的抑制;GA通过抑制DELLA,解除了对SOC1和LFY的抑制;随着植物年龄的增长通过miRNA156-miR172级联模式,解除了AP2-like对FT、AP1和SOC1的抑制等等;2) 植物最终实现开花转换,必然是多条途径共同作用,合力跨越“某个阈值”完成的,我们讨论的所有途径都可以调控和影响成花因子FT,但是ft突变体也只是表现晚花,那么到底哪个环节是“那根压死骆驼的稻草”?这是科学家们一直探索的动力。其实,由于物种、地域和进化的巨大差异,增加了植物开花机理的复杂性,所以,仍然需要进行多物种的深入研究才能最终完成这个具有挑战性的课题。
| [1] | Turck F, Fornara F, Coupland G. Regulation and identity of florigen: flowering locus T moves center stage. Annu Rev Plant Biol, 2008, 59: 573-594. |
| [2] | Valverde F, Mouradov A, Soppe W, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science, 2004, 303(5660): 1003-1006. |
| [3] |
Yang J, She SH. The family of Dof transcription factors in plants. Plant Physiol Comm, 2010, 46(3): 301-308 (in Chinese). 杨静, 沈世华. 植物Dof转录因子. 植物生理学通讯, 2010, 46(3): 301-308. |
| [4] | Sawa M, Nusinow DA, Kay SA, et al. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science, 2007, 318(5848): 261-265. |
| [5] | Imaizumi T, Schultz TF, Harmon FG, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science, 2005, 309(5732): 293-297. |
| [6] | Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science, 2012, 336(6084): 1045-1049. |
| [7] | Wang YZ, Gu XF, Yuan WY, et al. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell, 2014, 28(6): 727-736. |
| [8] | Ortiz-Marchena MI, Albi T, Lucas-Reina E, et al. Photoperiodic control of carbon distribution during the floral transition in Arabidopsis. Plant Cell, 2014, 26(2): 565-584. |
| [9] | Seo PJ, Ryu J, Kang SK, et al. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J, 2011, 65(3): 418-429. |
| [10] | Fornara F, Panigrahi KCS, Gissot L, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell, 2009, 17(1): 75-86. |
| [11] | Song YH, Estrada DA, Johnson RS, et al. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA, 2014, 111(49): 17672-17677. |
| [12] | Liu LJ, Zhang YC, Li QH, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell, 2008, 20(2): 292-306. |
| [13] | Kojima S, Takahashi Y, Kobayashi Y, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol, 2002, 43(10): 1096-1105. |
| [14] | Ben-Naim O, Eshed R, Parnis A, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J, 2006, 46(3): 462-476. |
| [15] | Lifschitz E, Eshed Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J Exp Bot, 2006, 57(13): 3405-3414. |
| [16] | Gyllenstrand N, Clapham D, Källman T, et al. A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol, 2007, 144(1): 248-257. |
| [17] | Bohlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science, 2006, 312(5776): 1040-1043. |
| [18] | Taylor A, Massiah AJ, Thomas B. Conservation of Arabidopsis thaliana photoperiodic flowering time genes in onion (Allium cepa L.). Plant Cell Physiol, 2010, 51(10): 1638-1647. |
| [19] | Navarro C, Abelenda JA, Cruz-Oró E, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature, 2011, 478(7367): 119-122. |
| [20] | Iwamoto M, Higo K, Takano M. Circadian clock-and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ, 2009, 32(5): 592-603. |
| [21] | Li DJ, Yang CH, Li XB, et al. Functional characterization of rice OsDof12. Planta, 2009, 229(6): 1159-1169. |
| [22] |
Yang J, Liu YP, Liu Y, et al. Progress in molecular biology of Jatropha curcas. Chin J Biotech, 2012, 28(6): 671-683 (in Chinese). 杨静, 刘永平, 刘蕴, 等. 麻疯树分子生物学研究进展. 生物工程学报, 2012, 28(6): 671-683. |
| [23] | Yang J, Yang MF, Zhang WP, et al. A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci, 2011, 181(6): 667-674. |
| [24] | Yang J, Yang MF, Wang D, et al. JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol Plant, 2010, 139(3): 324-334. |
| [25] | Kim DH, Sung S. Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book, 2014, 12: e0171. |
| [26] | Searle I, He YH, Turck F, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling inArabidopsis. Genes Dev, 2006, 20(7): 898-912. |
| [27] | Johanson U, West J, Lister C, et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 2000, 290(5490): 344-347. |
| [28] | Levy YY, Mesnage S, Mylne JS, et al. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science, 2002, 297(5579): 243-246. |
| [29] | Gendall AR, Levy YY, Wilson A, et al. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell, 2001, 107(4): 525-535. |
| [30] | Bastow R, Mylne JS, Lister C, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature, 2004, 427(6970): 164-167. |
| [31] | Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature, 2004, 427(6970): 159-164. |
| [32] | Gu XF, Le C, Wang YZ, et al. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun, 2013, 4: 1947. |
| [33] | Swiezewski S, Liu FQ, Magusin A, et al. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature, 2009, 462(7274): 799-802. |
| [34] | Liu FQ, Marquardt S, Lister C, et al. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science, 2010, 327(5961): 94-97. |
| [35] | Wang ZW, Wu Z, Raitskin O, et al. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci USA, 2014, 111(20): 7468-7473. |
| [36] | Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science, 2011, 331 (6013): 76-79. |
| [37] | Csorba T, Questa JI, Sun QW, et al. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA, 2014, 111(45): 16160-16165. |
| [38] | Reeves PA, He YH, Schmitz RJ, et al. Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics, 2007, 176(1): 295-307. |
| [39] | Kim SY, Park BS, Kwon SJ, et al. Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L.: ssp. pekinensis). Plant Cell Rep, 2007, 26(3): 327-336. |
| [40] | Locascio A, Lucchin M, Varotto S. Characterization of a MADS FLOWERING LOCUS C-LIKE (MFL) sequence in Cichorium intybus: a comparative study of CiMFL and AtFLC reveals homologies and divergences in gene function. New Phytol, 2009, 182(3): 630-643. |
| [41] | Shitsukawa N, Ikari C, Shimada S, et al. The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst, 2007, 82(2): 167-170. |
| [42] | Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends Plant Sci, 2009, 14(10): 563-573. |
| [43] | Yan LL, Loukoianov A, Blechl A, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science, 2004, 303(5664): 1640-1644. |
| [44] | Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet, 2008, 40(6): 761-767. |
| [45] | Yan L, Fu D, Li C, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA, 2006, 103(51): 19581-19586. |
| [46] | He YH. Chromatin regulation of flowering. Trends Plant Sci, 2012, 17 (9): 556-562. |
| [47] | Hornyik C, Terzi LC, Simpson GG. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell, 2010, 18(2): 203-213. |
| [48] | He YH, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science, 2003, 302(5651): 1751-1754. |
| [49] | Kim HJ, Hyun Y, Park JY, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet, 2004, 36(2): 167-171. |
| [50] | Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol, 2008, 59: 225-251. |
| [51] | Wilson RN, Heckman JW, Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol, 1992, 100(1): 403-408. |
| [52] | Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature, 2005, 437(7059): 693-698. |
| [53] | Murase K, Hirano Y, Sun TP, et al. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature, 2008, 456(7221): 459-463. |
| [54] | Davière JM, Achard P. Gibberellin signaling in plants. Development, 2013, 140(6): 1147-1151. |
| [55] | Yu S, Galvão VC, Zhang YC, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell, 2012, 24(8): 3320-3332. |
| [56] | Yamaguchi N, Winter CM, Wu MF, et al. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science, 2014, 344(6184): 638-641. |
| [57] | Goldberg-Moeller R, Shalom L, Shlizerman L, et al. Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci, 2013, 198: 46-57. |
| [58] | Ghosh A, Chikara J, Chaudhary DR, et al. Paclobutrazol arrests vegetative growth and unveils unexpressed yield potential of Jatropha curcas. J Plant Growth Regul, 2010, 29(3): 307-315. |
| [59] | Boden SA, Weiss D, Ross JJ, et al. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell, 2014, 26(4): 1557-1569. |
| [60] | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell, 2009, 136(2): 215-233. |
| [61] | Allen RS, Li JY, Stahle MI, et al. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA, 2007, 104(41): 16371-16376. |
| [62] | Alonso-Peral MM, Li JY, Li YJ, et al. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol, 2010, 154(2): 757-771. |
| [63] | Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 2009, 138(4): 738-749. |
| [64] | Wu G, Park MY, Conway SR, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 2009, 138(4): 750-759. |
| [65] | Wang JW. Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot, 2014, 65(17): 4723-4730. |
| [66] | Xie KB, Wu CQ, Xiong LZ. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol, 2006, 142(1): 280-293. |
| [67] | Zhou CM, Zhang TQ, Wang X, et al. Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science, 2013, 340(6136): 1097-1100. |
| [68] | Wang JW, Park MY, Wang LJ, et al. miRNA control of vegetative phase change in trees. PLoS Genet, 2011, 7(2): e1002012. |
| [69] | Zhang XH, Zou Z, Zhang JH, et al. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett, 2011, 585(2): 435-439. |
| [70] | Chuck GS, Tobias C, Sun L, et al. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci USA, 2011, 108(42): 17550-17555. |
| [71] | Riese M, Höhmann S, Saedler H, et al. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene, 2007, 401(1/2): 28-37. |
| [72] | Yant L, Mathieu J, Dinh TT, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell, 2010, 22(7): 2156-2170. |
| [73] | Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot, 2011, 62(2): 487-495. |
| [74] | Jung JH, Seo YH, Seo PJ, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell, 2007, 19(9): 2736-2748. |
| [75] | Jung JH, Seo PJ, Ahn JH, et al. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J Biol Chem, 2012, 287(19): 16007-16016. |
| [76] | Li D, Liu C, Shen LS, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell, 2008, 15(1): 110-120. |
| [77] | Andrés F, Porri A, Torti S, et al. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA, 2014, 111(26): 2760-2769. |
| [78] | Cho HJ, Kim JJ, Lee JH, et al. Short vegetative phase (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett, 2012, 586(16): 2332-2337. |
| [79] | Yu S, Cao L, Zhou CM, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife, 2013, 2: e00269. |
| [80] | Yang L, Xu M, Koo Y, et al. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife, 2013, 2: e00260. |
 2015, Vol. 31
2015, Vol. 31