中国科学院微生物研究所、中国微生物学会主办
文章信息
- 胡茂志, 潘志明, 杨芸, 孟闯, 张磊, 耿士忠, 焦新安
- Maozhi Hu, Zhiming Pan, Yun Yang, Chuang Meng, Lei Zhang, Shizhong Geng, Xin'an Jiao
- 嵌合鞭毛蛋白在大肠杆菌BL21(DE3) 和沙门菌SL5928中的表达与组装分析
- Expression and assembly of chimeric flagellins in Escherichia coli BL21(DE3) and Salmonella SL5928
- 生物工程学报, 2017, 33(8): 1335-1342
- Chinese Journal of Biotechnology, 2017, 33(8): 1335-1342
- 10.13345/j.cjb.160368
-
文章历史
- Received: October 3, 2016
- Accepted: March 9, 2017
2 扬州大学 江苏省人兽共患病学重点实验室,江苏 扬州 225009
2 Jiangsu Key Laboratory of Zoonosis, Yangzhou University, Yangzhou 225009, Jiangsu, China
Bacterial flagellum is the motility device of bacterium and its elongated filament is composed primarily of many subunits of flagellin [1]. The N-and C-terminal regions of the Salmonella typhimurium phase 1 flagellin, encoded by the fliCi gene, have been reported to be highly conserved helix structure across different bacterial species to maintain the integrity of the filament structure [2], whereas the middle regions are hypervariable regions (HVR). During bacterial flagellar filament assembly, flagellin has to be exported from the cytoplasm to the tip of the growing filament and undergoes a process from unfolding to folding state [3-4]. Because the folding and assembly process is essential for flagellin to be expressed on the bacterial surface [5], the recombinant flagellin can be acquired as unfolding form within the cytoplasm (monomer) [6-7] or folding form showing on the surface of bacteria (polymer) [5, 8-9].
Bacterial flagellin is a potent trigger of host innate immune responses in eukaryotes [10-11]. Two forms of flagellin have been found to show different ascendency in the immune response against exogenous antigen, although they have all been reported to show adjuvanticity [12-13]. Besides, the residues of flagellin which are involved in Toll-like receptor-5 signaling and NLRC4 (NLR family, CARD domain containing 4) inflammasome response are hidden in the flagellum and only monomer is accessible for recognition by host cells [14-17]. Hence the folding and assembly process of flagellin may influence their biological function. It is reported that flagella combined with exogenous antigen can significantly promote the release of cytokine, when compared with flagella alone [12]. Thus, for the flagellin-specific response assay, the present of exogenous antigen is more practical for the vaccine development than flagellin alone.
Replacement of part of the HVR of flagellin by exogenous antigen is a potentially effective strategy for the vaccine development [18]. The monomeric flagellin expressed in Escherichia coli and the polymeric form directly isolated from Salmonella is usually used in the vaccine development [8, 13, 19-20]. Reports have shown that the linkage of flagellin with the ectodomain of matrix 2 protein of influenza A virus (M2e) is a good vaccine candidate [20-21]. In this experiment, in order to analyze the assembly and biological activities of two forms of flagellin, two tandem M2e genes were designed to replace the DNA sequence (607–684 bp) of the fliCi gene in HVR and the chimeric genes were expressed in E. coli and Salmonella to produce the monomer and polymer chimeric flagellin, respectively. Then, the protein conformation, polymerization and IL-1β release in the early stage of immunization were analyzed.
1 Materials and methods 1.1 Mice and bacteriaTen six-week-old female C57BL/6 mice were obtained from the Comparative Medicine Center of Yangzhou University, China. This study was carried out in accordance with the regulations set forward by the Chinese Ministry of Science and Technology. The protocol was approved by the Committee on the Ethics of Animal Experiments of Yangzhou University (Permit number: 2007-0005). All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering. The recombinant plasmid pET-fliC containing fliCi gene of S. typhimurium LT2 in the BamHⅠ/XhoⅠ sites of pET30a+ was constructed by Dr. Hui Zhang [18]. E. coli BL21(DE3), S. typhimurium LB5000 and aflagellar S. dublin SL5928 were used in the following cloning procedures. The wild-type flagellin of S. typhimurium SL7207 was used as a control.
1.2 Construction of recombinant plasmidsThe DNA fragment (5′-GATACTACGATTGCTTTAGACAATAGTACTTCCCTGCTGACCGAAGTTGAAACCCCGACCCGTAACGAATGGGAATGCCGTTGCTCCGATTCCTCCGATGGCGGCGGCTGCTCCCTGCTGACCGAAGTTGAAACCCCGACCCGTAACGAATGGGAATGCCGTTGCTCCGATTCCTCCGATAAATATTACGCCAAAGTTACCGTTACGGGG-3′) was synthesized by GenScript Company (Nanjing, China). In this sequence, two M2e genes (4–72 bp) were connected with a DNA sequence, 5′-GGCGG CGGCTGC-3′, to yield the M2e2 gene (underlined). At the two ends of the M2e2 gene, a partial HVR sequence of fliCi gene was included for overlap polymerase chain reaction (PCR) procedures. The 5′-terminal region of the fliCi gene, fliC-AC, was amplified from pET-fliC using the following primers: fliC-forward-A and fliC-reverse-C. The 3′-terminal region, fliC-DB, was amplified using the following primers: fliC-forward-D and fliC-reverse-B. The fliC-AC-M2e2 fragment was amplified using the following primers: fliC-forward-A and M2E2-reverse from fliC-AC and M2e2. The chimeric fliC/M2e2 gene was amplified using the following primers: fliC-forward-A and fliC-reverse-B from fliC-AC-M2e2 and fliC-DB (Table 1; Fig. 1). The PCR product was cloned into the BamHⅠ/XhoⅠ sites of pET30a+ to yield the recombinant plasmid, pET-fliC/M2e2.
| Primer name | DNA sequence (5′-3′) |
| fliC-forward-A | AGGGATCCATGGCACAAGTCATTAA |
| fliC-reverse-C | AGTACTATTGTCTAAAGCAATCG |
| fliC-forward-D | AAATATTACGCCAAAGTTACCG |
| fliC-reverse-B | TGCTCGAGTTAACGCAGTAAAGAGA |
| M2E2-reverse | CCCCGTAACGGTAACTTTGGCG |

|
| Figure 1 Schematic construction of the chimeric gene fliC/M2e2. |
| |
The plasmid pET-fliC/M2e2 was transformed into BL21(DE3). The recombinant bacteria, BL21 (DE3)(pET-fliC/M2e2), were cultured in LB agar plates containing 50 μg/mL kanamycin overnight at 37 ℃ and the kanamycin-resistant colonies were then amplified by culturing with 1 mmol/L isopropyl-β-D-thiogalactopyranoside (IPTG). The recombinant bacterium BL21(DE3)(pET) was used as a negative control. Affinity purification of His-tagged monomeric chimeric flagellin (mfliC/M) was performed with a nickel bead column (Qiagen, Valencia, CA) according to the manufacturer's instructions after cell lysis by ultrasonication.
1.4 Preparation of polymeric chimeric flagellin expressed in Salmonella SL5928The plasmid pET-fliC/M2e2 was electroporated into LB5000 and then transferred to SL5928 by P22HTint transduction, as previously described [18]. Polymeric chimeric flagellin (pfliC/M) in the surface of kanamycin-resistant recombinant SL5928(fliC/M2e2) was extracted by acid treatment followed by ammonium sulfate precipitation, as previously described [13].
1.5 Transmission electron microscopyThe growth of chimeric flagella in the surface of bacteria and purified proteins were observed by Tecnai-12 transmission electronic microscopy (Philips, Holland) after negative staining.
1.6 SDS-PAGE and Western blottingThe expression and purity of chimeric flagellin were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred onto nitrocellulose and treated with murine anti-flagellin polyclonal antibody for Western blotting assay.
1.7 Circular dichroismTo investigate the secondary structure of purified flagellin, pfliC/M and mfliC/M, the far-ultraviolet (UV) circular dichroism (CD) spectra were recorded using a J-810 spectropolarimeter (Jasco, Tokyo, Japan) using 0.2 cm path length cuvettes at 20 ℃. The concentration of all samples used was 50 μg/mL in 10 mmol/L phosphate buffer saline (PBS), pH 7.4. The CD spectrum of PBS was used as baseline. The mean spectra of three repeated scans in the range between 190 nm and 250 nm, corrected by subtraction of the buffer signal, were obtained by taking points every 0.1 nm with 1 nm slit width.
1.8 Dynamic light scatteringTo further investigate the polymerization of mfliC/M in solution, the dynamic light scattering (DLS) measurements for pfliC/M and mfliC/M were performed with an EEN 3690 Malvern Zetasizer (Malvern, Worcestershire, UK) at 20 ℃. The concentration of all samples used was adjusted to 50 μg/mL in PBS (pH 7.4).
1.9 IL-1β releasePrimary peritoneal exudate cells (PECs) of C57BL/6 mice were harvested by washing the peritoneal cavity with RPMI 1640 medium containing 10% fetal bovine serum (1640-FBS, Gibco, Carlsbad, CA), as previously described (Kumari and Saxena, 2011). Cells were plated in 96-well plates at 2×104 cells/well and incubated for 3 h at 37 ℃ with 1 μg/mL E. coli lipopolysaccharide (LPS). The supernatants were removed and the cells were washed with 1640-FBS. pfliC/M and mfliC/M were then transfected respectively using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and incubated for 3 h (Simon and Samuel, 2008). The non-transfected cells were used as controls. The secretion of IL-1β in supernatant was detected using a mouse enzyme-linked immunosorbent assay (ELISA) set (Biosciences, PharMingen, San Diego, CA).
1.10 Statistical analysisWithin each experiment, three to four replicate assays were conducted for each treatment. All statistical analyses were performed by Student's t-test using SPSS software (Version 13.0 for Windows, Chicago, IL). A value of P≤0.05 was considered to be significant.
2 Results and discussion 2.1 Identification of two forms of chimeric flagellinIn the filament assembly of bacterial flagellum, the unfolding subunit protein flagellin has to be transported via a narrow channel, and then fold before being assembled into the growing filament [3-4]. The transport process is accompanied with the change of protein conformation. In this study, in order to express different forms of chimeric flagellin, the recombinant plasmid pET-fliC/M2e2 containing the chimeric gene fliC/M2e2 was designed to express as monomeric or polymeric flagellin using different expression systems, respectively.
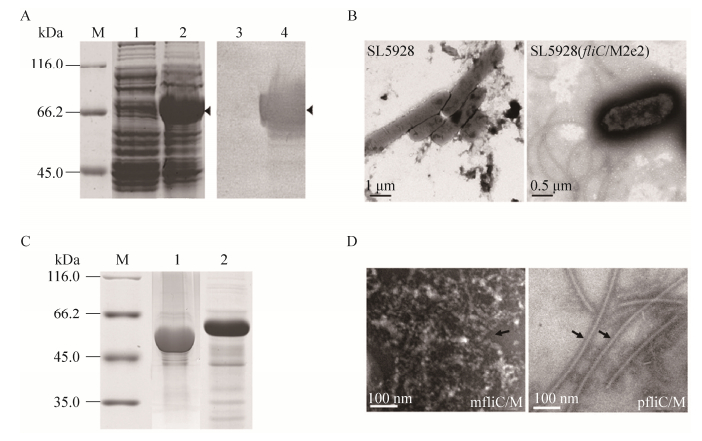
For the expression of monomeric chimeric flagellin, the SDS-PAGE and Western blotting results indicated that mfliC/M was successfully expressed in recombinant bacteria BL21(DE3) (pET-fliC/M2e2) when compared to BL21(DE3)(pET) (Fig. 2A). No flagellum was found on the surface of recombinant E. coli. For the expression of polymeric chimeric flagellin, polymeric flagella were observed on the bacterial surface of SL5928(fliC/M2e2) when compared to SL5928 (Fig. 2B). The molecular mass of pfliC/M and mfliC/M were about 50 kDa and 60 kDa, respectively (Fig. 2C). After purified, the polymeric pfliC/M showed a filament shape similar to wild-type flagellin and was significantly different from monomeric mfliC/M by transmission electronic microscope (Fig. 2D).
|
| Figure 2 Generation of chimeric flagellin pfliC/M (polymer) and mfliC/M (monomer). (A) SDS-PAGE (lane 1, 2) and Western blotting (lane 3, 4) analysis of recombinant bacteria induced with IPTG. M: standard molecular weight markers; lane 1 and 3: BL21(DE3)(pET); lane 2 and 4: BL21(DE3)(pET-fliC/M2e2). (B) Transmission electron microscopy observation of SL5928 and SL5928(fliC/M2e2) after negative staining. (C) SDS-PAGE of purified proteins. M: standard molecular weight markers; lane 1: pfliC/M; lane 2: mfliC/M. (D) Transmission electron microscopy observation of purified mfliC/M and pfliC/M after negative staining. |
| |
In BL21(DE3), the 6×His-tag was fusion expressed with fliC/M2e2 for purification of monermeric mfliC/M. While in SL5928, the fliC/M2e2 gene was integrated into the chromosome of SL5928 through homologous recombination [18]. In this study, the molecular weight between these two forms of chimeric flagellin was different. One reason may be due to the 6×His-tag in mfliC/M, besides post-translational modification.
2.2 Circular dichroism spectra and polymerization of two forms of chimeric flagellinThe folding and assembly of polymeric flagellin is one of the major difference compared with monomeric flagellin. CD spectroscopy is a common method for the analysis of protein structures in solution [22-23]. CD spectra between 190 and 250 nm are usually used to analyze the secondary structure of protein [23]. Moreover, the polymerization of proteins in solution can be determined by the size distribution by volume.
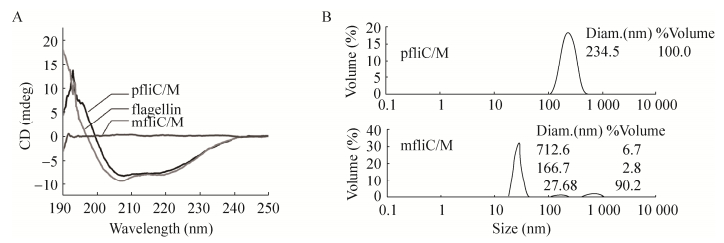
The far-UV CD measurements indicated that significantly different CD spectra between 190 and 250 nm were found between monomeric mfliC/M and polymeric pfliC/M. The CD signal of pfliC/M was stronger than that of mfliC/M. Similar CD spectra were observed between pfliC/M and wild-type polymeric flagellin of S. typhimurium SL7207 (Fig. 3A). The CD spectra of pfliC/M showed two negative peaks at 208 nm and 222 nm. This indicated that the helix conformation is its major secondary structure.
|
| Figure 3 Circular dichroism (A) and dynamic light scattering (B) assay of pfliC/M and mfliC/M. Results represent one of four independent experiments. |
| |
The polymerization of proteins in solution measured by DLS indicated that the mean dynamic radium of pfliC/M was larger than that of mfliC/M, though a few polymers were present in the mfliC/M solution (Fig. 3B). This indicated that the polymerization degree of pfliC/M was higher than that of mfliC/M.
The significant difference in transmission electronic microscope observation, CD spectra and polymerization between mfliC/M and pfliC/M may be due to the process of assembly. The terminal regions of flagellin are conserved helix structure to maintain the integrity of the filament structure [2]. The secondary structure of pfliC/M in this study showed typical helix conformation confirmed this point. Whereas only low-level polymerization of mfliC/M were found in vitro, thus we suppose that the polymerization of pfliC/M in vivo is a complex process. The middle region of flagellin is a HVR varying greatly in size and composition in different strains, and the insertion of exogenous antigen into HVR does not interfere with the assembly and export function [1, 3-4]. This study also verified that replacement of part of the HVR by M2e did not interfere with the conformation of polymeric flagellin.
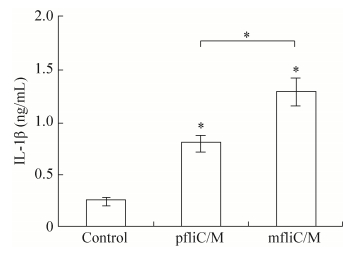
2.3 IL-1β release of PECs after transfectionLPS does not activate NLRC4 [24], but can induce the production of pro-IL-1β [25]. The C-terminal amino acid sequence of flagellin, which was hidden in the inner of filament, can be sensed by NLRC4 in macrophages and then trigger the maturation and release of IL-1β [4, 17]. Thus, the level of IL-1β secreted by macrophages can be used to indicate the activation of the NLRC4 signaling pathway. Report has shown that the secretion of IL-1β can be stimulated after transfection of monomeric flagellin to PECs [26-27]. In the study, three hours after transfection, the IL-1β assay indicated that pfliC/M and mfliC/M could significantly induce the secretion of IL-1β in PECs compared with the untransfected group (P < 0.05). The secretion level of IL-1β triggered by pfliC/M was lower than that for mfliC/M (P < 0.05) (Fig. 4). The relatively higher level of IL-1β secretion induced by mfliC/M may be due to the direct recognition by NLRC4 in the early stage, whereas polymeric flagellin must dissociate to release the monomeric form of flagellin first. While the detailed mechanism need to be further studied.
|
| Figure 4 IL-1β secretion. Primary peritoneal exudate cells (2×104/well) from C57BL/6 mice were incubated with E. coli LPS (1 μg/mL) for 3 h to induce the production of pro-IL-1β. Then, pfliC/M or mfliC/M was transfected using Lipofectamine 2 000 and incubated for 3 h. The concentration of IL-1β in supernatant was detected by ELISA. Non-transfected cells were used as control. These experiments were repeated three times. |
| |
Flagellin can be recognized by the innate immune system and has been used as adjuvant in the vaccine development [28]. The different expression and assembly assay of chimeric flagellin found in this study will be beneficial for their use in the future.
| [1] |
Malapaka RRV, Adebayo LO, Tripp BC. A deletion variant study of the functional role of the Salmonella flagellin hypervariable domain region in motility. J Mol Biol, 2007, 365(4): 1102-1116. DOI:10.1016/j.jmb.2006.10.054 |
| [2] |
Muskotál A, Király R, Sebestyén A, et al. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett, 2006, 580(16): 3916-3920. DOI:10.1016/j.febslet.2006.06.024 |
| [3] |
Chng C, Kitao A. Thermal unfolding simulations of bacterial flagellin: insight into its refolding before assembly. Biophys J, 2008, 94(10): 3858-3871. DOI:10.1529/biophysj.107.123927 |
| [4] |
Chng CP, Kitao A. Mechanical unfolding of bacterial flagellar filament protein by molecular dynamics simulation. J Mol Graph Modell, 2010, 28(6): 548-554. DOI:10.1016/j.jmgm.2009.11.007 |
| [5] |
Cuadros C, Lopez-Hernandez FJ, Dominguez AL, et al. Flagellin fusion proteins as adjuvants or vaccines induce specific immune responses. Infect Immun, 2004, 72(5): 2810-2816. DOI:10.1128/IAI.72.5.2810-2816.2004 |
| [6] |
Qian F, Li MM, Chen Y, et al. Salmonella flagellin acted as an effective fusion partner for expression of Plasmodium falciparum surface protein 25 in Escherichia coli. Hum Vaccin Immunother, 2016, 12(9): 2362-2364. DOI:10.1080/21645515.2016.1171443 |
| [7] |
Song L, Xiong D, Kang XL, et al. An avian influenza A (H7N9) virus vaccine candidate based on the fusion protein of hemagglutinin globular head and Salmonella typhimurium flagellin. BMC Biotechnol, 2015, 15(1): 79. DOI:10.1186/s12896-015-0195-z |
| [8] |
Braga CJM, Massis LM, Sbrogio-Almeida ME, et al. CD8+ T cell adjuvant effects of Salmonella FliCd flagellin in live vaccine vectors or as purified protein. Vaccine, 2010, 28(5): 1373-1382. DOI:10.1016/j.vaccine.2009.11.003 |
| [9] |
Le Moigne V, Robreau G, Mahana W. Flagellin as a good carrier and potent adjuvant for Th1 response: study of mice immune response to the p27 (Rv2108) Mycobacterium tuberculosis antigen. Mol Immunol, 2008, 45(9): 2499-2507. DOI:10.1016/j.molimm.2008.01.005 |
| [10] |
Brunner R, Jensen-Jarolim E, Pali-Schöll. The ABC of clinical and experimental adjuvants—a brief overview. Immunol Lett, 2010, 128(1): 29-35. DOI:10.1016/j.imlet.2009.10.005 |
| [11] |
Mizel SB, Graff AH, Sriranganathan N, et al. Flagellin-F1-V fusion protein is an effective plague vaccine in mice and two species of nonhuman primates. Clin Vaccine Immunol, 2009, 16(1): 21-28. DOI:10.1128/CVI.00333-08 |
| [12] |
Rochon M, Römling U. Flagellin in combination with curli fimbriae elicits an immune response in the gastrointestinal epithelial cell line HT-29. Microbes Infect, 2006, 8(8): 2027-2033. DOI:10.1016/j.micinf.2006.03.003 |
| [13] |
Skountzou I, del Pilar Martin M, Wang BZ, et al. Salmonella flagellins are potent adjuvants for intranasally administered whole inactivated influenza vaccine. Vaccine, 2010, 28(24): 4103-4112. DOI:10.1016/j.vaccine.2009.07.058 |
| [14] |
Zhang L, Pan ZM, Kang XL, et al. Amino acids 89-96 of Salmonella typhimurium flagellin represent the major domain responsible for TLR5-independent adjuvanticity in the humoral immune response. Cell Mol Immunol, 2015, 12(5): 625-632. DOI:10.1038/cmi.2014.76 |
| [15] |
Forstnerič V, Ivičak-Kocjan K, Ljubetič A, et al. Distinctive recognition of flagellin by human and mouse Toll-like receptor 5. PLoS ONE, 2016, 11(7): e0158894. DOI:10.1371/journal.pone.0158894 |
| [16] |
Voogdt CGP, Bouwman LI, Kik MJL, et al. Reptile Toll-like receptor 5 unveils adaptive evolution of bacterial flagellin recognition. Sci Rep, 2016, 6: 19046. DOI:10.1038/srep19046 |
| [17] |
Zhang H, Liu L, Wen K, et al. Chimeric flagellin expressed by Salmonella typhimurium induces an ESAT-6-specific Th1-type immune response and CTL effects following intranasal immunization. Cell Mol Immunol, 2011, 8(6): 496-501. DOI:10.1038/cmi.2011.24 |
| [18] |
Bargieri DY, Leite JA, Lopes SCP, et al. Immunogenic properties of a recombinant fusion protein containing the C-terminal 19 kDa of Plasmodium falciparum merozoite surface protein-1 and the innate immunity agonist FliC flagellin of Salmonella typhimurium. Vaccine, 2010, 28(16): 2818-2826. DOI:10.1016/j.vaccine.2010.02.004 |
| [19] |
Du LY, Zhou YS, Jiang SB. Research and development of universal influenza vaccines. Microbes Infect, 2010, 12(4): 280-286. DOI:10.1016/j.micinf.2010.01.001 |
| [20] |
Huleatt JW, Nakaar V, Desai P, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine, 2008, 26(2): 201-214. DOI:10.1016/j.vaccine.2007.10.062 |
| [21] |
Ceaglio N, Etcheverrigaray M, Kratje R, et al. Influence of carbohydrates on the stability and structure of a hyperglycosylated human interferon alpha mutein. Biochimie, 2010, 92(8): 971-978. DOI:10.1016/j.biochi.2010.04.004 |
| [22] |
Meersman F, Atilgan C, Miles AJ, et al. Consistent picture of the reversible thermal unfolding of hen egg-white lysozyme from experiment and molecular dynamics. Biophys J, 2010, 99(7): 2255-2263. DOI:10.1016/j.bpj.2010.07.060 |
| [23] |
Furukawa Y, Inoue Y, Sakaguchi A, et al. Structural stability of flagellin subunit affects the rate of flagellin export in the absence of FliS chaperone. Mol Microbiol, 2016, 102(3): 405-416. DOI:10.1111/mmi.2016.102.issue-3 |
| [24] |
Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1β in mouse macrophage. J Immunol, 2008, 180(11): 7147-7157. DOI:10.4049/jimmunol.180.11.7147 |
| [25] |
Simon R, Samuel CE. Interleukin-1 beta secretion is activated comparably by FliC and FljB flagellins but differentially by wild-type and DNA adenine methylase-deficient Salmonella. J Interf Cytokine Res, 2010, 28(11): 661-666. |
| [26] |
Zhao Y, Yang JL, Shi JJ, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type Ⅲ secretion apparatus. Nature, 2011, 477(7366): 596-600. DOI:10.1038/nature10510 |
| [27] |
Matusiak M, Van Opdenbosch N, Vande Walle L, et al. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc Natl Acad Sci USA, 2015, 112(5): 1541-1546. DOI:10.1073/pnas.1417945112 |
| [28] |
Xiao XX, Zhang Y, Liu JX, et al. Immunoenhancement with flagellin as an adjuvant to whole-killed rabies vaccine in mice. Arch Virol, 2016, 161(3): 685-691. DOI:10.1007/s00705-015-2704-8 |
 2017, Vol. 33
2017, Vol. 33




