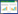中国科学院微生物研究所、中国微生物学会主办
文章信息
- 于珊, 马旅雁
- Shan Yu, Luyan Ma
- 铜绿假单胞菌铁摄取与生物被膜形成研究进展
- Iron uptake and biofilm formation in Pseudomonas aeruginosa
- 生物工程学报, 2017, 33(9): 1489-1512
- Chinese Journal of Biotechnology, 2017, 33(9): 1489-1512
- 10.13345/j.cjb.170140
-
文章历史
- Received: April 5, 2017
- Accepted: May 8, 2017
2 中国科学院微生物研究所, 北京 100101
2 Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
生物被膜(Biofilm)是指单细胞微生物通过粘附于介质表面,分泌胞外多聚基质(胞外多糖、蛋白、DNA等),将其自身包绕其中而成的膜样微生物细胞聚集物[1]。生物被膜能够保护微生物在不利环境中得以存活并在条件有利时散播种子细胞去占领新的地盘。生物被膜可以在多种介质表面形成,在自然界、工业环境和医院等场所都普遍存在。这种固着的聚集状态的微生物,其生理指标与游离的菌细胞相比存在很大的差异[2-3]。在临床环境中,细菌生物被膜形成后会对抗生素的抗性大大提高并产生多种选择性的表型突变,因而可引起多种难以清除的慢性感染。对生物被膜在基因和分子水平的形成机制上的深入了解可为防治生物被膜引起的临床感染和问题提供新的解决策略。
生物被膜的胞外多聚物(EPS)是由多糖、胞外DNA (eDNA)和蛋白质等组成的混合物(图 1A)。EPS可以形成网络,并像分子胶水一样将细菌细胞网罗在一起(图 1B)。这种基质网对维持生物被膜的整体结构和抗生素的抗性起到很大作用[4-5]。揭示EPS在生物被膜形成中的作用可为设计对抗生物被膜的靶向分子提供有益的思路。
条件致病菌铜绿假单胞菌具有极强的生存能力,极易形成生物被膜,是研究生物被膜的模式生物之一。该菌极易感染免疫力较弱的患者,如患代谢性疾病、血液病和恶性肿瘤的患者,以及术后或某些治疗后的患者(如癌症、大面积烧伤、HIV等),导致极高的发病率和致死率。铜绿假单胞菌可在囊性纤维化(Cystic fibrosis,CF)患者的肺部定殖并导致难以根除的慢性感染。已证实这种慢性感染难以根除的原因之一就是因为铜绿假单胞菌可在患者的肺部形成生物被膜。铜绿假单胞菌在不同的营养条件下可形成不同形态的生物被膜[8]。例如,在以葡萄糖为碳源的限制性培养基中,铜绿假单胞菌PAO1的生物被膜发育周期可分为5个典型阶段(图 2)。

|
| 图 2 铜绿假单胞菌生物被膜的典型发育周期[7]. 1:可逆性吸附期:浮游微生物附着至载体表面的初期,附着于载体的微生物也可以回到单细胞浮游状态(Ⅰ);2:不可逆吸附期(Ⅱ);3:生物被膜形成初期(Ⅲ);4:生物被膜成熟期(Ⅳ);5:“种子散播期” (Ⅴ). (Ⅰ–Ⅴ)在生物被膜发育的每个阶段,Psl多糖是如何包被生物被膜中的细菌及其在相应时期的变化(Psl多糖由红色荧光标记,菌体由绿色荧光标记) Figure 2 Scheme of biofilm development in P. aeruginosa[7]. Selected images showed how the matrix of Psl polysaccharide (red fluorescence) enmeshes bacterial cells (green fluorescence) within bacterial communities during biofilm development. 1: initial attachment (Ⅰ); 2: irreversible attachment (Ⅱ); 3: microcolony formation (Ⅲ); 4: biofilm maturation (Ⅳ); 5: biofilm dispersion (Ⅴ). |
| |
本文介绍了铜绿假单胞菌生物被膜形成调控机制及其基质网的各组分的近期研究进展,并进一步详细阐述了其铁摄取的主要系统与策略。深入了解铜绿假单胞菌生物被膜的形成机制和生存策略,可为解决铜绿假单胞菌引起的问题与感染提供途径与思路。
1 铜绿假单胞菌生物被膜基质网铜绿假单胞菌在肺部的定殖是肺囊性纤维化病(CF)患者发病和死亡主要原因。铜绿假单胞菌造成的感染通常与形成抗生素抗性极强的生物被膜有关,菌细胞被包裹在基质网中形成微菌落聚集体。即使长期使用大量的抗生素进行治疗,感染仍然会持续存在,非常难以清除[9-10]。EPS基质网所起的重要作用与菌株的遗传背景、营养条件和生物被膜所处的不同发展阶段相关[11]。普遍接受的观点认为,EPS可以作为生物被膜的结构骨架并在不利的环境下为菌细胞提供保护屏障[12-13]。在铜绿假单胞菌中,至少存在3种胞外多糖在维持生物被膜的结构和抗生素抗性方面起着重要作用[5, 7, 12, 14-16]。与生物被膜形成、发展及功能相关的基质网组成部分主要有胞外多糖(Psl多糖、Pel多糖和褐藻多糖)、胞外DNA、胞外蛋白以及菌体表面的一些蛋白附属结构,如纤毛、四型菌毛(T4P)及鞭毛。
1.1 Psl多糖2004年3个不同的研究组分别鉴定出了Psl的多糖合成位点[14-15, 17]。Psl多糖是由15个共转录的基因(pslA-pslO, PA2231–PA2245) 组成的基因簇所编码的一系列蛋白共同合成的。Psl多糖对于菌体的初始吸附、生物被膜结构的维持以及提供细胞与细胞间、细胞与介质表面的相互作用至关重要[7, 18-20]。相关研究进一步表明psl合成基因簇的15个基因中的前11个是合成Psl多糖所必需的[21],铜绿假单胞菌PA14由于pslA-pslD基因的缺失导致其丧失了合成Psl多糖的能力[22]。早先有研究表明Psl多糖富含半乳糖、甘露糖及少量的葡萄糖和木糖[18]。最近有研究进一步证实了Psl多糖是由D-甘露糖、D-葡萄糖和L-鼠李糖形成的四糖重复单位组成的[21]。此外,在基因及生化水平对单个的psl基因,如pslA、pslB和pslD的功能研究揭示出这些基因对Psl多糖的合成及生物被膜的形成都是非常关键的[23-25]。
由于Psl多糖在铜绿假单胞菌生物被膜的形成中起着非常重要的作用,因而对其在生物被膜形成中的具体功能和角色已经有了许多非常广泛深入的研究[20]。首先,Psl多糖的过量生产可以增强菌细胞之间、菌细胞与介质表面间的黏附作用,提示Psl多糖在吸附中的重要功能对起始和维持生物被膜的结构非常关键[19, 21]。后来Ma及其同事用荧光标记的凝集素进一步直接观察到了Psl多糖在生物被膜不同发育阶段的形态和结构(图 2),而且发现了Psl多糖是以一种双螺旋的形态锚定在菌细胞表面并通过这种锚定方式有效地促进了菌细胞之间的相互作用,并以此完成早期生物被膜及其基质网的形成和组装。在生物被膜形成后期,Psl多糖主要聚集在立体的微菌落结构的外围。这种定位方式为生物被膜提供了结构支撑并便于之后的种子散播[7]。除此之外,对Psl多糖的染色观察表明Psl可以形成一种纤维状的网络将菌细胞网罗在生物被膜中。近来,有研究发现Psl多糖基质网是通过依赖四型菌毛的迁移策略形成的,T4P介导的细菌迁移导致了Psl纤维基质网的形成,缺失了菌毛的突变株丧失了形成Psl纤维结构的能力并且导致生物被膜的生物量下降[26]。Zhao及其同事也同时证明铜绿假单胞菌通过在介质表面的迁移留下Psl多糖踪迹,进一步影响后续菌细胞的表面运动并最终导致生物被膜形成的起始[27]。以上这些最新发现都扩展了对Psl多糖的生物学理解。
令人惊讶的是,Psl多糖还具有信号功能,可通过刺激两个二鸟苷酸环化酶SiaD和SadC来升高胞间第二信使分子c-di-GMP的水平,形成一个独特的正反馈调节回路最终提高Psl多糖的产量[28]。
此外,Psl多糖在赋予菌体抗原性及保护菌细胞免受宿主免疫系统攻击等方面也有作用。Psl多糖可通过增强菌细胞与上皮细胞的接触直接刺激NF-κB的活性并促进鞭毛介导的促炎信号[29]。Mishra等发现铜绿假单胞菌的Psl多糖可通过限制补体介导的调理作用减弱中性白细胞的吞噬以及氧化应激反应的发生[30]。此类研究清晰地表明Psl多糖对宿主内环境中的菌体生存具有重要意义,并提供了将Psl多糖作为靶点清除生物被膜从而治疗相关慢性感染的直接依据。
有研究证实Psl多糖在细菌抗生素耐药性的产生中也发挥作用。最近有研究揭示Psl多糖可以促进细菌对生物被膜抑制剂聚山梨醇酯80 (Polysorbate 80) 的抗性[31]。Yang等报道生物被膜中依赖Psl多糖形成的微菌落结构使铜绿假单胞菌对抗生素的处理变得不敏感[20]。通过用荧光标记的抗生素观察到胞外基质网可通过限制妥布霉素的渗透保护生物被膜中的菌细胞免遭抗生素的杀伤,且这种对抗生素的隔离作用主要发生在生物被膜的外周区域[32]。然而,没有明确证据能直接证实生物被膜基质网对这种保护和相互作用的关键功能。此外,另一个研究组报道在生物被膜的形成起始阶段Psl多糖作为第一道防线来抵御阳离子或阴离子类抗生素的攻击,且这种由Psl多糖介导的保护作用可以扩展到混合培养生物被膜中的大肠杆菌和金黄色葡萄球菌[33]。
总之,Psl多糖不仅可作为生物被膜发育过程中的结构骨架,还可以作为一种信号分子存在最终导致生物被膜的形成。这种正反馈回路代表了一种铜绿假单胞菌在不同环境中生存定殖的进化策略。此外,Psl多糖还可作为屏障来抵御宿主免疫和抗生素的杀伤。
1.2 Pel多糖Pel多糖是由pelA-pelF (PA3058-PA3064) 组成的pel合成基因簇编码的蛋白合成。Pel多糖是一种富含葡萄糖且对纤维素处理敏感的胞外多聚物[34]。pel合成基因簇在革兰氏阴性菌中比较保守[35]。这部分基因的缺失会导致细菌在静置培养条件下无法形成气液交界面的菌膜,此外,Pel多糖在介质表面形成牢固的生物被膜过程中也发挥作用[14]。有趣的是,缺失菌毛的铜绿假单胞菌PAK的pel突变株,其在固体介质表面的初始吸附能力大大减弱了,提示Pel多糖可能在其他一些黏附因子如四型菌毛缺失时可以提供代偿作用[7]。然而,Pel多糖在其他铜绿假单胞菌菌株的吸附过程中的明确作用仍需进一步深入的研究。此外,Pel多糖通过维持细胞间的相互粘连在菌株PA14形成的生物被膜中为菌细胞提供主要的骨架支撑,并通过增强生物被膜对氨基糖甙类抗生素的抗性为细菌提供保护[16]。进一步对Pel功能的研究揭示Pel多糖可以和其他类型的EPS共同作用来促进铜绿假单胞菌的生物被膜发育[20]。
1.3 褐藻多糖褐藻多糖(Alginate)是CF患者肺部临床分离株产生的主要胞外多糖[36]。典型黏液型表型的产生原因是由于菌株生产了大量的褐藻多糖,这种多糖形成的生物被膜胞外基质网可以保护铜绿假单胞菌免受肺部不利环境的影响。然而,这种多糖对于体外非黏性生物被膜的形成并不是必需的[37]。褐藻多糖在保护和维持生物被膜结构稳定等方面起着重要作用——有保水及储存营养物质的功能[4]。有趣的是,近来的研究发现,黏性菌株生物被膜的形成也依赖Psl多糖的产生[38-39]。褐藻多糖还有维持细胞存活和免疫逃避的功能[40]。褐藻多糖的大量生产可以增强菌细胞对抗生素和调离吞噬作用的抵抗能力[41-42]。这种多糖在体内可以清除由中性粒细胞和巨噬细胞产生的自由基[43]。Bragonizi等也曾报道褐藻多糖的大量合成并没有增强铜绿假单胞菌在小鼠感染模型中长久生存能力,这也许是由于感染过程中菌株从黏液型到非黏液型的表型转变有关[44]。
1.4 胞外DNA胞外DNA (eDNA)是铜绿假单胞菌生物被膜基质网中的另一重要组分[17, 45-46]。eDNA可能源于基因组DNA,在生物被膜中提供细胞间的相互联结。对生物被膜中的eDNA进行染色发现其主要大量存在于蘑菇状微菌落的柄部[45]。此外,在生物被膜微菌落中也存在着菌细胞的自溶现象,但目前尚不明确eDNA是否来源于菌细胞自溶释放出的基因组DNA[47]。关于eDNA的来源目前主要的几个假设有:来源于由前噬菌体裂解的亚群体、由菌细胞直接分泌及由小的外膜囊泡产生等[48]。
与生物被膜基质网中的其他组分相同,eDNA对生物被膜的形成也具有多重作用,有研究证实外源添加DNaseⅠ可以降解较为“年轻”的生物被膜,此外eDNA也与抗生素抗性的产生有关[49]。有趣的是,eDNA还可在饥饿状态时作为菌细胞的营养来源[50],近来有研究表明,铜绿假单胞菌产生一种胞外脱氧核糖核酸酶(PA3909) 可利用eDNA作为营养来吸收利用,这进一步扩展了我们对于eDNA在铜绿假单胞菌生物被膜形成过程中作用的理解[51]。此外,eDNA还可以与坏死的中性粒细胞释放出的F-肌动蛋白形成束状物,这些束状物可以促进铜绿假单胞菌生物被膜的形成起始[52]。eDNA作为铜绿假单胞菌生物被膜中的主要促炎因子,通过CpG和TLR9-依赖的机制激活中性粒细胞[53]。进一步的研究发现eDNA可以通过维持细胞间的粘连和排列来调节生物被膜边缘部位先锋细胞群的运动,进而促进由颤动作用介导的生物被膜扩张[54]。
1.5 胞外蛋白及菌体表面蛋白附属结构除胞外多糖和胞外DNA外,胞外蛋白与四型菌毛、鞭毛和纤毛等蛋白多聚体细胞附属物也是组成生物被膜基质网的成分。这些细胞附属物主要作为黏附因子和结构支撑为铜绿假单胞菌生物被膜的形成提供辅助作用[55]。
鞭毛可以介导铜绿假单胞菌的游动(Swimming)和丛集运动(Swarming),还可以作为黏附因子为细胞和介质表面的初始接触提供关键助力[56]。四型菌毛(T4P)是一种线状的运动器官,采用一种“伸展-握紧-撤回”的机制形成铜绿假单胞菌的颤动(Twitching)能力[57]。四型菌毛在铜绿假单胞菌微菌落的典型“蘑菇冠”结构的形成中起着重要作用[56]。如上所述,T4P介导的菌体运动可以帮助Psl多糖纤维丝状网的形成[26],而另有研究认为铜绿假单胞菌的鞭毛和菌毛并不是初始吸附或形成生物被膜所必需的[58],即鞭毛或菌毛介导的生物被膜形成可能是依条件和营养而异的。近来,有研究证实一个新鉴定的粘附素CdrA是铜绿假单胞菌EPS基质网中的关键组分蛋白,通过直接与Psl多糖作用介导菌体的自聚集并增强生物被膜的稳定性[59]。
铜绿假单胞菌的纤毛类附属物由“分子伴侣-引导蛋白(Chaperon-Usher)”机制组装而成,其对生物被膜的形成亦具有促进作用[60]。在铜绿假单胞菌PAO1中,鉴定出至少4个纤毛系统(CupA、B、C和D)并分别具有不同的组织形式和功能[61]。有研究表明Cup纤毛有利于生物被膜的形成起始,特别是在介导细胞间互作和微菌落的形成过程中起着关键作用[18, 62]。
2 铜绿假单胞菌生物被膜形成的调控生物被膜的形成受控于多种复杂的细菌调控体系之下,包括群体感应系统(Qurum sensing,QS)及参与调节胞外多糖合成的双组分调控系统等[63]。这些调控系统的缺失会导致生物被膜的结构、形态及保护能力的变化。现对铜绿假单胞菌与生物被膜形成相关的关键调控系统进行简要概述。
2.1 群体感应系统与生物被膜形成群体感应是细菌群体的一种细胞间的交流方式,通过生产及接受能扩散的信号分子感应菌细胞的群体密度,并通过这种机制调控毒力因子的产生、运动力的变化及生物被膜的形成[64-65]。铜绿假单胞菌具有两种主要的QS系统(las和rhl),分别通过合成酶LasI、RhlI合成两种自诱导信号分子:N-3-氧十二烷酰高丝氨酸内酯(3-oxo-C12-HSL)和N-丁酰高丝氨酸内酯(C4-HSL)[65]。信号分子与相应的受体蛋白(转录因子LasR和RhlR)结合后激活一系列基因(包括lasI和rhlI在内)的转录。此外,铜绿假单胞菌还有第3个QS系统——PQS系统,依赖喹诺酮信号分子,与酰基高丝氨酸内酯(AHLs)信号分子之间存在着复杂的交互调控关系[65] (图 3)。
Davies等已证明了las系统在铜绿假单胞菌生物被膜形成和成熟过程中的作用[2]。与野生型相比,las系统缺失株形成更扁平、均一化的生物被膜,且受到十二烷基硫酸钠的处理后更易从介质表面剥离,但las系统在生物被膜形成中的具体功能尚不明确。而后Gilbert等报道QS转录因子LasR可以结合在psl操纵子的启动子区——表明QS系统可以调控psl基因簇的表达[67]。rhl系统亦被证实可以通过促进Pel多糖的合成参与铜绿假单胞菌生物被膜形成:在rhl系统缺失株中pel操纵子的转录水平明显降低[68]。而PQS系统与生物被膜发育过程中eDNA的释放有关,pqsA突变株形成的生物被膜中的eDNA含量比野生型生物被膜中的更少[45, 69]。综上所述,以上3种QS系统均在铜绿假单胞菌生物被膜的发育过程中起着较为重要的调控作用。
不能忽略的是,QS系统还可通过调控铜绿假单胞菌的丛集运动、颤动及鼠李糖脂、凝集素的产量来间接影响生物被膜的形成过程。铜绿假单胞菌的丛集运动能力是由鞭毛、菌毛和胞间接触共同介导的一种复杂的表面运动能力[70-71]。rhl系统可以通过调节丛集运动参与早期铜绿假单胞菌生物被膜的形成[72]。铜绿假单胞菌在促进丛集运动的培养基(如以谷氨酸或琥珀酸为碳源的生长培养基)中形成扁平均一的生物被膜,而在丧失丛集运动的情况下形成融合样细胞聚集状态的生物被膜[8]。颤动能力是一种由极生四型菌毛介导的一种细菌运动[73],已知其在铁缺乏的限制性培养基中受到rhl系统的调控[74]。颤动能力是铜绿假单胞菌在生物被膜形成过程中将单层细胞组装成微菌落的立体结构所必需的[56]。
鼠李糖脂,除了作为一种表面活性剂和铜绿假单胞菌的毒力因子[75],可受到rhl系统的调控并在生物被膜的形成中起多种作用[76]:1) 参与生物被膜微菌落的形成[77];2) 通过干扰细胞-细胞、细胞-介质表面间的相互作用阻碍菌细胞定殖并维持生物被膜中通道结构的开放[78];3) 通过促进3D蘑菇状结构的形成帮助铜绿假单胞菌生物被膜发育[77];4) 高产鼠李糖脂的突变株更易从生物被膜中脱离,因此鼠李糖脂的过量表达可以促进铜绿假单胞菌生物被膜的瓦解[79]。最后,铜绿假单胞菌的细胞毒力因子——凝集素LecA和LecB,也参与生物被膜的形成,LecA和LecB的突变株只能形成比野生型更薄的生物被膜[80-81],而LecA和LecB的表达也是受到rhl系统调控的[82]。
2.2 GacS/GacA和RetS/LadS双组分系统对生物被膜的调控在铜绿假单胞菌基因组中的60个双组分调控系统中[83],GacS/GacA系统参与调控QS系统并调节多种毒力因子的产生和生物被膜的形成[84]。Gac系统由一个跨膜感应激酶GacS和一个配对反应调节蛋白GacA组成,GacS可通过自磷酸化转移一个磷酸基给GacA,磷酸化的GacA可上调两个小RNA——RsmZ和RsmY的表达,RsmZ和RsmY可结合在一种小RNA结合蛋白RsmA上,而RsmA在转录后水平抑制psl合成簇(pslA-L)的转录[85-86](图 4)。GacS/GacA系统还对AHL系统有调控作用:可通过使游离的RsmA失活解除其对C4-HSL和3-oxo-C12-HSL合成的抑制作用,从而通过las和rhl系统调控胞外毒力因子的产生[87-88]。
有报道称混合传感组氨酸激酶RetS可抑制生物被膜的形成[90-91],而另一个组氨酸激酶LadS对RetS有拮抗作用[92]。与野生型菌株PAO1相比,retS突变株可形成结构化程度更高的生物被膜[90];而天然缺乏ladS基因的菌株PA14与ladS过表达株相比,所形成的生物被膜更薄[93]。有研究表明,RetS和LadS与GacS/GacA系统之间存在着交互作用:即可通过调节GacS的磷酸化状态来抑制或促进GacA的磷酸化[93-94]。有趣的是,GacS/GacA和RetS/LadS系统还参与调控铜绿假单胞菌从急性感染到慢性感染的表型转变过程[90]。
2.3 环二鸟苷酸(C-di-GMP)依赖的多糖合成与生物被膜的形成铜绿假单胞菌胞外多糖的合成依赖于胞内环二鸟苷酸(c-di-GMP)的浓度[95-96],c-di-GMP是细菌中广泛存在的一种胞内第二信使[97]。在菌细胞内,两分子的鸟苷三磷酸(GTP)在二鸟苷酸环化酶的催化下合成一分子的c-di-GMP,后者又可在底物专一性的磷酸二酯酶的催化下降解为两分子的鸟苷酸(GMP) [97](图 5)。
高浓度的c-di-GMP可通过与合成褐藻多糖和Pel多糖所必需的Alg44和PelD蛋白结合分别促进这两种多糖的合成[96-98]。然而,这种c-di-GMP调控糖前体多聚化的具体机制尚不明确。反过来,低浓度的c-di-GMP可通过加强鞭毛运动促进菌细胞的游动和解聚[96]。
3 铜绿假单胞菌的铁摄取系统铁离子是地球上绝大多数生物生存必不可少的元素之一,生物体内的很多酶催化反应都需要铁离子的参与。然而,由于氧的存在,环境中的大部分铁离子都以三价铁的形式存在,很难溶解于水溶液中。因此,环境中的铁看起来很多,实际上大部分都是铁的化合物,自由铁的浓度很低,满足不了微生物的生长需求。此外,由于宿主铁大多被亚铁血红素分子及转铁蛋白、乳铁蛋白之类的循环蛋白所结合,因而病原微生物在宿主体内也面临着严峻的铁摄取问题[99-100],细菌为了利用铁来维持自身生长并完成各种生理功能,进化出了一系列能够螯合、摄取铁的系统,而这些系统对细菌毒力因子的释放和生物被膜的形成均起着重要的调控作用。
铜绿假单胞菌可以通过如下不同策略来获取铁元素:1) 生产Fe3+螯合分子,即铁载体:荧光嗜铁素(Pyoverdine)和鳌铁蛋白(Pyochelin),并通过TonB依赖的受体(TonB-dependent receptors,TBDR)摄入三价铁-载体复合物。2) 摄入外源铁载体复合物(非铜绿假单胞菌本身产生的铁载体)。3) 从宿主产生的血红素蛋白摄入血红素分子。4) 利用吩嗪(Phenazine)和Fe2+专一性摄取系统(Feo系统)在胞外将Fe3+还原为Fe2+并加以吸收利用。5) 在宿主体内,依据造成感染的不同类型(急性感染或慢性感染),铜绿假单胞菌能够采取不同的铁摄取策略在不造成过多能量消耗的前提下满足自身的铁需求。
3.1 铜绿假单胞菌所产铁载体及其介导的铁摄取系统铁载体是一类能够以高亲和力专一性螯合Fe3+的低分子量的分泌型分子,能够依赖由TonB周质膜蛋白所提供的能量被特异性受体识别[101-105]。铁载体依据其与铁所形成的复合物的形式,可分为不同的种类,如酚盐铁载体、苯酚铁载体、异羟肟酸铁载体、羧化铁载体及混合铁载体。
3.2 荧光嗜铁素:引发急性感染所必需的高亲和力铁载体铜绿假单胞菌荧光嗜铁素是由一个肽链和一个发色团组成的混合型铁载体[106-108]。荧光嗜铁素是能发荧光的一些假单胞菌(如荧光假单胞菌、恶臭假单胞菌、丁香假单胞菌和铜绿假单胞菌)的标志性特点,其在环境铁浓度过低时被产生出来[106-108]。荧光嗜铁素与铁的亲和力极高,可将铁从转铁蛋白中螯合出来,且荧光嗜铁素的产生是引起小鼠烧伤模型和肺病模型的感染所必需的[106, 109-113]。同样地,TonB蛋白的突变株丧失了在小鼠模型中的致病力[111]。在Imperi等的研究中,筛选到了一些通过FDA认证的抑制铜绿假单胞菌荧光嗜铁素合成的化合物,其中一个化合物——抗真菌药物氟胞嘧啶对多株铜绿假单胞菌产生的不同类型的荧光嗜铁素均有明显的抑制作用,且这种抑制作用是通过作用于σ因子PvdS来实现的,而PvdS是荧光嗜铁素合成基因簇转录所必需的调节因子[113]。荧光嗜铁素不仅是一种铁载体,还是一种信号分子,可诱导两种胞外毒力因子蛋白酶PrpL和外毒素A的产生[108, 114-117] (图 6)。此外,尽管荧光嗜铁素是铜绿假单胞菌引起急性感染所必需的因素,其也在较厚的成熟生物被膜形成过程中发挥作用[74, 118-119]。
3.3 鳌铁蛋白:一种低亲和力铁载体鳌铁蛋白是所有铜绿单胞菌均产生的一种铁载体,但其对铁的亲和力较荧光嗜铁素低[121-123]。鳌铁蛋白合成所需的基因数比荧光嗜铁素所需基因数更少[124],且有研究表明铜绿假单胞菌优先合成鳌铁蛋白,在环境铁浓度非常低的情况下再切换为荧光嗜铁素的合成[125]。鳌铁蛋白-铁复合物可以进行氧化还原循环并引起宿主的氧化损伤和炎症反应,尤其是在铜绿假单胞菌的另一种胞外产物绿脓菌素也存在的情况下[126-128]。在慢性感染过程中,如在CF患者肺部,鳌铁蛋白的产生可引起持续的炎症反应并导致组织损伤[129]。在CF痰培养基中,铜绿假单胞菌的鳌铁蛋白合成量增加[130]。
3.4 亚铁血红素摄取系统铜绿假单胞菌可以通过Has和Phu两套系统从血红蛋白中摄取亚铁血红素[131] (图 7)。亚铁血红素无法以自由游离形式存在,因为强疏水性使其与膜相结合并在膜上进行非酶促氧化还原反应[132],因此亚铁血红素必须从血红蛋白或血液结合素上提取得到。在铜绿假单胞菌的Phu系统中,亚铁血红素是从一种外膜TBDR中提取到的,而在Has系统中,亚铁血红素先由血红蛋白摄取,然后这种“血红素载体-血红素”复合物再被另一种TBDR——HasR识别[133-135]。在细胞周质中,血红素与周质结合蛋白结合后通过ABC转运系统运输到胞内。而在ABC转运系统中,血红素先与血红素分子伴侣PhuS结合,然后被运输给血红素氧化酶HemO,进一步将血红素降解为胆绿素、CO和Fe2+[136-138]。对Has或Phu系统的一个单突变不会影响血红素摄取,而对两个系统的双突变则会导致铜绿假单胞菌完全丧失将血红素作为铁源的摄取能力[131]。
铜绿假单胞菌至少具有30个以上的基因用来编码TBDRs,而其中大多数都参与铁与载体复合物的摄入[139-141]。不同的TBDRs可以分为两类:简单的TBDR和TonB依赖的转换子,即TBDT[142]。以TBDT系统中的“铁-荧光嗜铁素”复合物受体FpvA为例,FpvA可以通过与一个作为抗σ因子的膜蛋白相互作用来感受“铁-荧光嗜铁素”复合物的存在[142]。这种受体识别一旦发生就会使抗σ因子蛋白水解,从而释放出外ECFσ (Extracytoplasmic function sigma factor),ECFσ可与RNA聚合酶作用,从而引发下游基因转录自诱导反应[117, 140, 142-143]。大多数的铜绿假单胞菌(98%)都具有另一种“一型铁-荧光嗜铁素受体”——FpvB,这意味着几乎所有菌株都能利用这种荧光嗜铁素作为铁源[139, 144]。铜绿假单胞菌还可以通过各种不同的受体来摄取各种各样由其他细菌产生的“铁-载体复合物”,这种能够“偷取”其他细菌铁源的能力,可能赋予其在混合感染中较强的竞争优势[145]。
3.6 通过Feo系统与吩嗪摄入Fe2+与Fe3+不同,Fe2+具有水溶性,并存在于低pH值的厌氧或微厌氧环境中[146]。Fe2+可以扩散至外膜并通过在革兰氏阴性菌中广泛存在的FeoABC系统转运至细胞质[147] (图 8)。Fe2+通过由透性酶FeoB和FeoA、FeoC蛋白组成的转运系统继续被运输至细胞内部[147]。Fe2+的摄入可能与铜绿假单胞菌对微厌氧或厌氧环境的感受有关,如CF患者的肺部的生物被膜黏液环境[148-149]。
吩嗪类物质是一种由铜绿假单胞菌产生的次级代谢产物[150],1-羧基-吩嗪酸(PCA)是绿脓菌素的前体,绿脓菌素是铜绿假单胞菌所产生的一种蓝绿色特征性化合物,这两种吩嗪物质都具有氧化还原能力[150]。PCA及一些少量的绿脓菌素可将宿主蛋白结合的Fe3+还原为Fe2+,在生物被膜中Fe2+可经由Feo系统摄入[151]。近来有研究报道当病情恶化时,可在CF患者肺部检测到吩嗪和Fe2+的大量积累[152-153],而Fe2+和Fe3+两种离子的螯合剂联用可以大大削弱铜绿假单胞菌的生物被膜形成能力[153]。
3.7 铜绿假单胞菌铁平衡的调节机制与大多数细菌一样,Fur是假单胞菌的一个主要的铁调节蛋白,且在铜绿假单胞菌中,fur是生存必需基因[154]。大部分基因参与“铁-载体复合物”摄取的基因受到Fur的间接调控(图 9)。Fur调控一些ECFσ因子的表达,这些σ因子包括荧光嗜铁素生物合成基因pvdS、荧光嗜铁素复合物基因fpvA和其他一些编码TonB依赖型外源铁复合物受体的基因,它们是实现一些下游基因转录所必需的。铁浓度可作为信号通过ECFσ因子引发这些基因的表达。受体与“铁-载体复合物”的结合改变了受体蛋白N端与膜抗σ因子的相互作用,释放出ECFσ因子,使后者能够与核RNA聚合酶结合。铜绿假单胞菌还具有另一种铁调节机制,即通过AraC调节子来控制鳌铁蛋白的合成与铁摄取[155]。最后,两个小RNA——PrrF1和PrrF2可以作为mRNA反义转录后调节子参与Fur的调节,主要通过参与上调氧化还原应激、铁储存和中间代谢等Fe-Fur基因的表达[156-157]。
综上所述,铜绿假单胞菌可以从合成较低铁亲和力的鳌铁蛋白转为合成较高铁亲和力但更耗能量的荧光嗜铁素来摄取足够量的Fe3+[125]。一个典型的例子即发生在铜绿假单胞菌对CF患者肺部环境的适应过程中[129]。当铜绿假单胞菌开始侵染肺部时,可能产生荧光嗜铁素,但随其在肺部的进一步定殖,细菌开始由于某些原因合成鳌铁蛋白并引发强烈的炎症反应,进而导致组织损伤和包括血红蛋白和其他含铁蛋白等细胞内容物的释放[128]。尽管包括荧光嗜铁素在内的铁载体在CF患者的痰液样本中被检测到[159],但不能产生荧光嗜铁素的突变株具有更久的定殖能力[160-161],这意味着铜绿假单胞菌可以利用其他代偿性机制来满足自身的铁需求,例如可以从炎症反应中释放出的血红蛋白中摄取血红素,或者摄取经由吩嗪类物质,特别是PCA介导的氧化还原反应产生的Fe2+[151, 153, 161-162]。
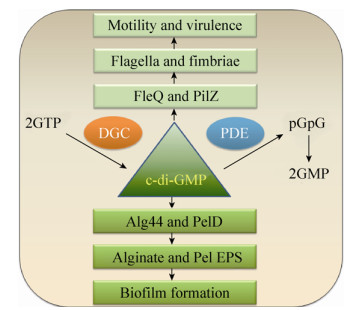
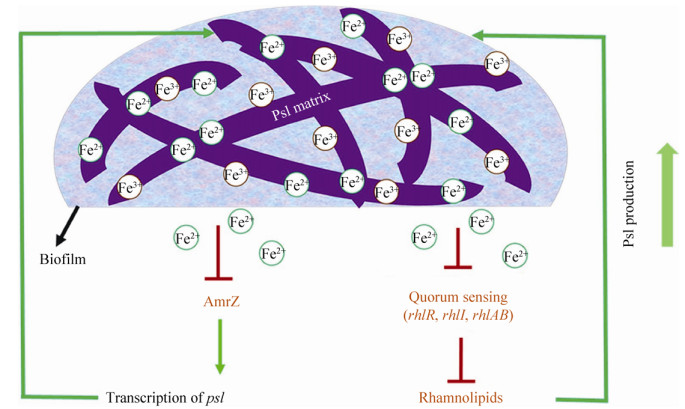
4 铁与生物被膜形成有研究发现外源添加铁螯合剂乳铁蛋白可对铜绿假单胞菌生物被膜的形成产生明显的抑制作用[163],而适宜的高铁浓度环境则促进其生物被膜的形成[164-165]。这表明环境铁可作为一种信号调节铜绿假单胞菌生物被膜的发育。与上述研究结论相一致,研究者还发现丧失了螯合环境铁能力的铜绿假单胞菌突变株不能形成正常的生物被膜[74, 118]。这种铁对铜绿假单胞菌生物被膜的调节效应可能是受到铁受体调节子Fur的调控而产生[141, 166]。近年来有研究报道高铁浓度可以抑制铜绿假单胞菌黏性菌株的主要胞外多糖——褐藻多糖的产生,使其生物被膜的形成能力降低[167]。最新研究发现,铜绿假单胞菌的胞外多糖Psl可直接结合环境中的自由铁,而高铁浓度信号通过抑制转录因子AmrZ降低鼠李糖脂的合成量,从而通过解除其对Psl多糖的糖前体竞争抑制而产生更多的Psl多糖,形成更大生物量的生物被膜(图 10),这是一种铜绿假单胞菌摄取铁的新策略[168]。
本文概述了铜绿假单胞菌生物被膜发展发育的主要过程及其系统调控网络,并特别介绍了铁摄取系统的工作原理及铁信号与其生物被膜形成之间的关系。进一步增强了人们对于微生物生物被膜发育过程以及环境适应性的认识,同时为生物被膜的防控提供了潜在的候选药物靶标。
| [1] | Karatan E, Watnick P. Signals, regulatory networks and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev, 2009, 73(2): 310–347. DOI: 10.1128/MMBR.00041-08 |
| [2] | Davies DG, Parsek MR, Pearson JP, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science, 1998, 280(5361): 295–298. DOI: 10.1126/science.280.5361.295 |
| [3] | Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol, 2004, 2(2): 95–108. DOI: 10.1038/nrmicro821 |
| [4] | Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology, 2001, 147(1): 3–9. DOI: 10.1099/00221287-147-1-3 |
| [5] | Branda SS, Vik Å, Friedman L, et al. Biofilms: the matrix revisited. Trends Microbiol, 2005, 13(1): 20–26. DOI: 10.1016/j.tim.2004.11.006 |
| [6] | Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol, 2010, 8(9): 623–633. |
| [7] | Ma LY, Conover M, Lu HP, et al. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog, 2009, 5(3): e1000354. DOI: 10.1371/journal.ppat.1000354 |
| [8] | Shrout JD, Chopp DL, Just CL, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol, 2006, 62(5): 1264–1277. DOI: 10.1111/mmi.2006.62.issue-5 |
| [9] | Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science, 1999, 284(5418): 1318–1322. DOI: 10.1126/science.284.5418.1318 |
| [10] | Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature, 2002, 416(6882): 740–743. DOI: 10.1038/416740a |
| [11] | Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the " house of biofilm cells". J Bacteriol, 2007, 189(22): 7945–7947. DOI: 10.1128/JB.00858-07 |
| [12] | Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol, 2007, 10(6): 644–648. DOI: 10.1016/j.mib.2007.09.010 |
| [13] | Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol, 2003, 57: 677–701. DOI: 10.1146/annurev.micro.57.030502.090720 |
| [14] | Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol, 2004, 186(14): 4457–4465. DOI: 10.1128/JB.186.14.4457-4465.2004 |
| [15] | Jackson KD, Starkey M, Kremer S, et al. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol, 2004, 186(14): 4466–4475. DOI: 10.1128/JB.186.14.4466-4475.2004 |
| [16] | Colvin KM, Gordon VD, Murakami K, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog, 2011, 7(1): e1001264. DOI: 10.1371/journal.ppat.1001264 |
| [17] | Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol, 2004, 186(14): 4449–4456. DOI: 10.1128/JB.186.14.4449-4456.2004 |
| [18] | Ruer S, Stender S, Filloux A, et al. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol, 2007, 189(9): 3547–3555. DOI: 10.1128/JB.00093-07 |
| [19] | Ma LY, Jackson KD, Landry RM, et al. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol, 2006, 188(23): 8213–8221. DOI: 10.1128/JB.01202-06 |
| [20] | Yang L, Hu YF, Liu Y, et al. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol, 2011, 13(7): 1705–1717. DOI: 10.1111/j.1462-2920.2011.02503.x |
| [21] | Byrd MS, Sadovskaya I, Vinogradov E, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol, 2009, 73(4): 622–638. DOI: 10.1111/mmi.2009.73.issue-4 |
| [22] | Winsor GL, Lam DKW, Fleming L, et al. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res, 2011, 39(S1). |
| [23] | Campisano A, Schroeder C, Schemionek M, et al. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl Environ Microbiol, 2006, 72(4): 3066–3068. DOI: 10.1128/AEM.72.4.3066-3068.2006 |
| [24] | Overhage J, Schemionek M, Webb JS, et al. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl Environ Microbiol, 2005, 71(8): 4407–4413. DOI: 10.1128/AEM.71.8.4407-4413.2005 |
| [25] | Lee HJ, Chang HY, Venkatesan N, et al. Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1. FEBS Lett, 2008, 582(23/24): 3479–3483. |
| [26] | Wang SW, Parsek MR, Wozniak DJ, et al. A spider web strategy of type Ⅳ pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ Microbiol, 2013, 15(8): 2238–2253. DOI: 10.1111/emi.2013.15.issue-8 |
| [27] | Zhao K, Tseng BS, Beckerman B, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature, 2013, 497(7449): 388–391. DOI: 10.1038/nature12155 |
| [28] | Irie Y, Borlee BR, O'Connor JR, et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA, 2012, 109(50): 20632–20636. DOI: 10.1073/pnas.1217993109 |
| [29] | Byrd MS, Pang B, Mishra M, et al. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-κB activation in A549 cells. mBio, 2010, 1(3): e00140–10. |
| [30] | Mishra M, Byrd MS, Sergeant S, et al. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol, 2012, 14(1): 95–106. DOI: 10.1111/cmi.2012.14.issue-1 |
| [31] | Zegans ME, Wozniak D, Griffin E, et al. Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob Agents Chemother, 2012, 56(8): 4112–4122. DOI: 10.1128/AAC.00373-12 |
| [32] | Tseng BS, Zhang W, Harrison JJ, et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol, 2013, 15(10): 2865–2878. |
| [33] | Billings N, Millan MR, Caldara M, et al. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog, 2013, 9(8): e1003526. DOI: 10.1371/journal.ppat.1003526 |
| [34] | Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol, 2004, 51(3): 675–690. |
| [35] | Vasseur P, Vallet-Gely I, Soscia C, et al. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology, 2005, 151(3): 985–997. DOI: 10.1099/mic.0.27410-0 |
| [36] | Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Mol Biol Rev, 1996, 60(3): 539–574. |
| [37] | Wozniak DJ, Wyckoff TJO, Starkey M, et al. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA, 2003, 100(13): 7907–7912. DOI: 10.1073/pnas.1231792100 |
| [38] | Ma LY, Wang SW, Wang D, et al. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol, 2012, 65(2): 377–380. DOI: 10.1111/j.1574-695X.2012.00934.x |
| [39] | Yang L, Wang HZ, Wu H, et al. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol Med Microbiol, 2012, 65(2): 366–376. DOI: 10.1111/j.1574-695X.2012.00936.x |
| [40] | Leid JG, Willson CJ, Shirtliff ME, et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol, 2005, 175(11): 7512–7518. DOI: 10.4049/jimmunol.175.11.7512 |
| [41] | Simpson JA, Smith SE, Dean RT. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. Microbiology, 1988, 134(1): 29–36. DOI: 10.1099/00221287-134-1-29 |
| [42] | Simpson JA, Smith SE, Dean RT. Alginate may accumulate in cystic fibrosis lung because the enzymatic and free radical capacities of phagocytic cells are inadequate for its degradation. Biochem Mol Biol Int, 1993, 30(6): 1021–1034. |
| [43] | Simpson JA, Smith SE, Dean RT. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med, 1989, 6(4): 347–353. DOI: 10.1016/0891-5849(89)90078-6 |
| [44] | Bragonzi A, Paroni M, Nonis A, et al. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med, 2009, 180(2): 138–145. DOI: 10.1164/rccm.200812-1943OC |
| [45] | Allesen-Holm M, Barken KB, Yang L, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol, 2006, 59(4): 1114–1128. DOI: 10.1111/mmi.2006.59.issue-4 |
| [46] | Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science, 2002, 295(5559): 1487. DOI: 10.1126/science.295.5559.1487 |
| [47] | Webb JS, Thompson LS, James S, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol, 2003, 185(15): 4585–4592. DOI: 10.1128/JB.185.15.4585-4592.2003 |
| [48] | Spoering AL, Gilmore MS. Quorum sensing and DNA release in bacterial biofilms. Curr Opin Microbiol, 2006, 9(2): 133–137. DOI: 10.1016/j.mib.2006.02.004 |
| [49] | Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog, 2008, 4(11): e1000213. DOI: 10.1371/journal.ppat.1000213 |
| [50] | Finkel SE, Kolter R. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol, 2001, 183(21): 6288–6293. DOI: 10.1128/JB.183.21.6288-6293.2001 |
| [51] | Mulcahy H, Charron-Mazenod L, Lewenza S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol, 2010, 12(6): 1621–1629. |
| [52] | Parks QM, Young RL, Poch KR, et al. Neutrophil enhancement of Pseudomonas aeruginosa biofilm development: human F-actin and DNA as targets for therapy. J Med Microbiol, 2009, 58(4): 492–502. DOI: 10.1099/jmm.0.005728-0 |
| [53] | Fuxman Bass JI, Russo DM, Gabelloni ML, et al. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J Immunol, 2010, 184(11): 6386–6395. DOI: 10.4049/jimmunol.0901640 |
| [54] | Gloag ES, Turnbull L, Huang A, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA, 2013, 110(28): 11541–11546. DOI: 10.1073/pnas.1218898110 |
| [55] | Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev, 2012, 36(4): 893–916. DOI: 10.1111/j.1574-6976.2011.00322.x |
| [56] | O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol, 1998, 30(2): 295–304. DOI: 10.1046/j.1365-2958.1998.01062.x |
| [57] | Skerker JM, Berg HC. Direct observation of extension and retraction of type Ⅳ pili. Proc Natl Acad Sci USA, 2001, 98(12): 6901–6904. DOI: 10.1073/pnas.121171698 |
| [58] | Klausen M, Heydorn A, Ragas P, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type Ⅳ pili mutants. Mol Microbiol, 2003, 48(6): 1511–1524. DOI: 10.1046/j.1365-2958.2003.03525.x |
| [59] | Borlee BR, Goldman AD, Murakami K, et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol, 2010, 75(4): 827–842. DOI: 10.1111/mmi.2010.75.issue-4 |
| [60] | Vallet I, Olson JW, Lory S, et al. The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA, 2001, 98(12): 6911–6916. DOI: 10.1073/pnas.111551898 |
| [61] | Giraud C, Bernard CS, Calderon V, et al. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol, 2011, 13(3): 666–683. DOI: 10.1111/emi.2011.13.issue-3 |
| [62] | Kulasekara HD, Ventre I, Kulasekara BR, et al. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol, 2005, 55(2): 368–380. |
| [63] | de Kievit TR. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol, 2009, 11(2): 279–288. DOI: 10.1111/emi.2009.11.issue-2 |
| [64] | Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol, 2005, 13(1): 27–33. DOI: 10.1016/j.tim.2004.11.007 |
| [65] | Jimenez PN, Koch G, Thompson JA, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev, 2012, 76(1): 46–65. DOI: 10.1128/MMBR.05007-11 |
| [66] | Lee J, Zhang LH. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell, 2015, 6(1): 26–41. DOI: 10.1007/s13238-014-0100-x |
| [67] | Gilbert KB, Kim TH, Gupta R, et al. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol, 2009, 73(6): 1072–1085. DOI: 10.1111/mmi.2009.73.issue-6 |
| [68] | Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol, 2007, 189(14): 5383–5386. DOI: 10.1128/JB.00137-07 |
| [69] | Yang L, Barken KB, Skindersoe ME, et al. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology, 2007, 153(5): 1318–1328. DOI: 10.1099/mic.0.2006/004911-0 |
| [70] | Fraser GM, Hughes C. Swarming motility. Curr Opin Microbiol, 1999, 2(6): 630–635. DOI: 10.1016/S1369-5274(99)00033-8 |
| [71] | Harshey RM. Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol, 1994, 13(3): 389–394. DOI: 10.1111/mmi.1994.13.issue-3 |
| [72] | Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev, 2004, 28(3): 261–289. DOI: 10.1016/j.femsre.2003.09.004 |
| [73] | Mattick JS. type Ⅳ pili and twitching motility. Annu Rev Microbiol, 2002, 56: 289–314. DOI: 10.1146/annurev.micro.56.012302.160938 |
| [74] | Patriquin GM, Banin E, Gilmour C, et al. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol, 2008, 190(2): 662–671. DOI: 10.1128/JB.01473-07 |
| [75] | Van Gennip M, Christensen LD, Alhede M, et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS, 2009, 117(7): 537–546. DOI: 10.1111/j.1600-0463.2009.02466.x |
| [76] | Dusane DH, Zinjarde SS, Venugopalan VP, et al. Quorum sensing: implications on Rhamnolipid biosurfactant production. Biotechnol Genet Eng Rev, 2010, 27(1): 159–184. DOI: 10.1080/02648725.2010.10648149 |
| [77] | Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol, 2007, 189(6): 2531–2539. DOI: 10.1128/JB.01515-06 |
| [78] | Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol, 2003, 185(3): 1027–1036. DOI: 10.1128/JB.185.3.1027-1036.2003 |
| [79] | Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol, 2005, 57(5): 1210–1223. DOI: 10.1111/j.1365-2958.2005.04743.x |
| [80] | Diggle SP, Stacey RE, Dodd C, et al. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol, 2006, 8(6): 1095–1104. DOI: 10.1111/emi.2006.8.issue-6 |
| [81] | Tielker D, Hacker S, Loris R, et al. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology, 2005, 151(5): 1313–1323. DOI: 10.1099/mic.0.27701-0 |
| [82] | Winzer K, Falconer C, Garber NC, et al. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol, 2000, 182(22): 6401–6411. DOI: 10.1128/JB.182.22.6401-6411.2000 |
| [83] | Rodrigue A, Quentin Y, Lazdunski A, et al. Cell signalling by oligosaccharides. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol, 2000, 8(11): 498–504. |
| [84] | Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol, 2001, 40(5): 1215–1226. DOI: 10.1046/j.1365-2958.2001.02469.x |
| [85] | Brencic A, McFarland KA, McManus HR, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol, 2009, 73(3): 434–445. DOI: 10.1111/mmi.2009.73.issue-3 |
| [86] | Irie Y, Starkey M, Edwards AN, et al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol, 2010, 78(1): 158–172. |
| [87] | Kay E, Humair B, Dénervaud V, et al. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol, 2006, 188(16): 6026–6033. DOI: 10.1128/JB.00409-06 |
| [88] | Reimmann C, Beyeler M, Latifi A, et al. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol, 1997, 24(2): 309–319. DOI: 10.1046/j.1365-2958.1997.3291701.x |
| [89] | Jimenez PN, Koch G, Thompson JA, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev, 2012, 76(1): 46–65. DOI: 10.1128/MMBR.05007-11 |
| [90] | Goodman AL, Kulasekara B, Rietsch A, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell, 2004, 7(5): 745–754. DOI: 10.1016/j.devcel.2004.08.020 |
| [91] | Kong W, Chen L, Zhao JQ, et al. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol Microbiol, 2013, 88(4): 784–797. DOI: 10.1111/mmi.2013.88.issue-4 |
| [92] | Ventre I, Goodman AL, Vallet-Gely I, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA, 2006, 103(1): 171–176. DOI: 10.1073/pnas.0507407103 |
| [93] | Goodman AL, Merighi M, Hyodo M, et al. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev, 2009, 23(2): 249–259. DOI: 10.1101/gad.1739009 |
| [94] | Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type Ⅵ secretion and virulence factors. J Bacteriol, 2010, 192(14): 3584–3596. DOI: 10.1128/JB.00114-10 |
| [95] | Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA, 2005, 102(40): 14422–14427. DOI: 10.1073/pnas.0507170102 |
| [96] | Merighi M, Lee VT, Hyodo M, et al. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol, 2007, 65(4): 876–895. DOI: 10.1111/mmi.2007.65.issue-4 |
| [97] | Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol, 2009, 7(4): 263–273. DOI: 10.1038/nrmicro2109 |
| [98] | Lee VT, Matewish JM, Kessler JL, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol, 2007, 65(6): 1474–1484. DOI: 10.1111/mmi.2007.65.issue-6 |
| [99] | Finkelstein RA, Sciortino CV, McIntosh MA. Role of iron in microbe-host interactions. Rev Infect Dis, 1983, 5(S4): S759–S777. |
| [100] | Cornelissen CN, Sparling PF. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol, 1994, 14(5): 843–850. DOI: 10.1111/mmi.1994.14.issue-5 |
| [101] | Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci, 1999, 24(3): 104–109. DOI: 10.1016/S0968-0004(99)01359-6 |
| [102] | Boukhalfa H, Crumbliss AL. Chemical aspects of siderophore mediated iron transport. Biometals, 2002, 15(4): 325–339. DOI: 10.1023/A:1020218608266 |
| [103] | Hider RC, Kong XL. Chemistry and biology of siderophores. Nat Prod Rep, 2010, 27(5): 637–657. DOI: 10.1039/b906679a |
| [104] | Schalk IJ, Mislin GLA, Brillet K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr Top Membr, 2012, 69: 37–66. DOI: 10.1016/B978-0-12-394390-3.00002-1 |
| [105] | Schalk IJ, Guillon L. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids, 2013, 44(5): 1267–1277. DOI: 10.1007/s00726-013-1468-2 |
| [106] | Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol, 2000, 174(3): 135–142. DOI: 10.1007/s002030000188 |
| [107] | Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol, 2003, 11(5): 195–200. DOI: 10.1016/S0966-842X(03)00076-3 |
| [108] | Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol, 2007, 15(1): 22–30. DOI: 10.1016/j.tim.2006.11.004 |
| [109] | Albrecht-Gary AM, Blanc S, Rochel N, et al. Bacterial iron transport: coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg Chem, 1994, 33(26): 6391–6402. DOI: 10.1021/ic00104a059 |
| [110] | Meyer JM, Neely A, Stintzi A, et al. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun, 1996, 64(2): 518–523. |
| [111] | Takase H, Nitanai H, Hoshino K, et al. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect Immun, 2000, 68(8): 4498–4504. DOI: 10.1128/IAI.68.8.4498-4504.2000 |
| [112] | Takase H, Nitanai H, Hoshino K, et al. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun, 2000, 68(4): 1834–1839. DOI: 10.1128/IAI.68.4.1834-1839.2000 |
| [113] | Imperi F, Massai F, Facchini M, et al. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc Natl Acad Sci USA, 2013, 110(18): 7458–7463. DOI: 10.1073/pnas.1222706110 |
| [114] | Lamont IL, Beare PA, Ochsner U, et al. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci USA, 2002, 99(10): 7072–7077. DOI: 10.1073/pnas.092016999 |
| [115] | Rédly GA, Poole K. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J Bacteriol, 2003, 185(4): 1261–1265. DOI: 10.1128/JB.185.4.1261-1265.2003 |
| [116] | Rédly GA, Poole K. FpvIR control of fpvA ferric pyoverdine receptor gene expression in Pseudomonas aeruginosa: demonstration of an interaction between FpvI and FpvR and identification of mutations in each compromising this interaction. J Bacteriol, 2005, 187(16): 5648–5657. DOI: 10.1128/JB.187.16.5648-5657.2005 |
| [117] | Cornelis P. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol, 2010, 86(6): 1637–1645. DOI: 10.1007/s00253-010-2550-2 |
| [118] | Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA, 2005, 102(31): 11076–11081. DOI: 10.1073/pnas.0504266102 |
| [119] | Glick R, Gilmour C, Tremblay J, et al. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol, 2010, 192(12): 2973–2980. DOI: 10.1128/JB.01601-09 |
| [120] | Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol, 2013, 3: 75. |
| [121] | Cox CD, Rinehart KL Jr, Moore ML, et al. Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc Natl Acad Sci USA, 1981, 78(7): 4256–4260. DOI: 10.1073/pnas.78.7.4256 |
| [122] | Ankenbauer RG, Toyokuni T, Staley A, et al. Synthesis and biological activity of pyochelin, a siderophore of Pseudomonas aeruginosa. J Bacteriol, 1988, 170(11): 5344–5351. DOI: 10.1128/jb.170.11.5344-5351.1988 |
| [123] | Brandel J, Humbert N, Elhabiri M, et al. Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(Ⅲ), copper(Ⅳ) and zinc(Ⅱ) complexes. Dalton Trans, 2012, 41(9): 2820–2834. DOI: 10.1039/c1dt11804h |
| [124] | Serino L, Reimmann C, Visca P, et al. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J Bacteriol, 1997, 179(1): 248–257. DOI: 10.1128/jb.179.1.248-257.1997 |
| [125] | Dumas Z, Ross-Gillespie A, Kümmerli R. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc Roy Soc B Biol Sci, 2013, 280(1764): 20131055. DOI: 10.1098/rspb.2013.1055 |
| [126] | Coffman TJ, Cox CD, Edeker BL, et al. Possible role of bacterial siderophores in inflammation. Iron bound to the Pseudomonas siderophore pyochelin can function as a hydroxyl radical catalyst. J Clin Invest, 1990, 86(4): 1030–1037. DOI: 10.1172/JCI114805 |
| [127] | Britigan BE, Roeder TL, Rasmussen GT, et al. Interaction of the Pseudomonas aeruginosa secretory products pyocyanin and pyochelin generates hydroxyl radical and causes synergistic damage to endothelial cells. Implications for Pseudomonas-associated tissue injury. J Clin Invest, 1992, 90(6): 2187–2196. DOI: 10.1172/JCI116104 |
| [128] | Britigan BE, Rasmussen GT, Cox CD. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun, 1997, 65(3): 1071–1076. |
| [129] | Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev, 2002, 15(2): 194–222. DOI: 10.1128/CMR.15.2.194-222.2002 |
| [130] | Hare NJ, Soe CZ, Rose B, et al. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J Proteome Res, 2012, 11(2): 776–795. DOI: 10.1021/pr200659h |
| [131] | Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology, 2000, 146(1): 185–198. DOI: 10.1099/00221287-146-1-185 |
| [132] | Wyckoff EE, Lopreato GF, Tipton KA, et al. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J Bacteriol, 2005, 187(16): 5658–5664. DOI: 10.1128/JB.187.16.5658-5664.2005 |
| [133] | Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol, 1998, 28(6): 1223–1234. DOI: 10.1046/j.1365-2958.1998.00885.x |
| [134] | Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol, 2004, 58: 611–647. DOI: 10.1146/annurev.micro.58.030603.123811 |
| [135] | Wandersman C, Delepelaire P. Haemophore functions revisited. Mol Microbiol, 2012, 85(4): 618–631. DOI: 10.1111/mmi.2012.85.issue-4 |
| [136] | Bhakta MN, Wilks A. The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the δ-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry, 2006, 45(38): 11642–11649. DOI: 10.1021/bi060980l |
| [137] | Barker KD, Barkovits K, Wilks A. Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J Biol Chem, 2012, 287(22): 18342–18350. DOI: 10.1074/jbc.M112.359265 |
| [138] | O'Neill MJ, Bhakta MN, Fleming KG, et al. Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc Natl Acad Sci USA, 2012, 109(15): 5639–5644. DOI: 10.1073/pnas.1121549109 |
| [139] | Bodilis J, Ghysels B, Osayande J, et al. Distribution and evolution of ferripyoverdine receptors in Pseudomonas aeruginosa. Environ Microbiol, 2009, 11(8): 2123–2135. DOI: 10.1111/emi.2009.11.issue-8 |
| [140] | Cornelis P, Bodilis J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ Microbiol Rep, 2009, 1(4): 256–262. DOI: 10.1111/emi4.2009.1.issue-4 |
| [141] | Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals, 2009, 22(1): 15–22. DOI: 10.1007/s10534-008-9193-0 |
| [142] | Hartney SL, Mazurier S, Kidarsa TA, et al. TonB-dependent outer-membrane proteins and siderophore utilization in Pseudomonas fluorescens Pf-5. Biometals, 2011, 24(2): 193–213. DOI: 10.1007/s10534-010-9385-2 |
| [143] | Mettrick KA, Lamont IL. Different roles for anti-sigma factors in siderophore signalling pathways of Pseudomonas aeruginosa. Mol Microbiol, 2009, 74(5): 1257–1271. DOI: 10.1111/mmi.2009.74.issue-5 |
| [144] | Ghysels B, Dieu BTM, Beatson SA, et al. FpvB, an alternative type Ⅰ ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology, 2004, 150(6): 1671–1680. DOI: 10.1099/mic.0.27035-0 |
| [145] | Traxler MF, Seyedsayamdost MR, Clardy J, et al. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol, 2012, 86(3): 628–644. DOI: 10.1111/mmi.12008 |
| [146] | Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev, 2003, 27(2/3): 215–237. |
| [147] | Cartron ML, Maddocks S, Gillingham P, et al. Feo -transport of ferrous iron into bacteria. Biometals, 2006, 19(2): 143–157. DOI: 10.1007/s10534-006-0003-2 |
| [148] | Worlitzsch D, Tarran R, Ulrich M, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest, 2002, 109(3): 317–325. DOI: 10.1172/JCI0213870 |
| [149] | Yoon SS, Hennigan RF, Hilliard GM, et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell, 2002, 3(4): 593–603. DOI: 10.1016/S1534-5807(02)00295-2 |
| [150] | Wang Y, Newman DK. Redox reactions of phenazine antibiotics with ferric (hydr) oxides and molecular oxygen. Environ Sci Technol, 2008, 42(7): 2380–2386. DOI: 10.1021/es702290a |
| [151] | Wang Y, Wilks JC, Danhorn T, et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol, 2011, 193(14): 3606–3617. DOI: 10.1128/JB.00396-11 |
| [152] | Hunter RC, Klepac-Ceraj V, Lorenzi MM, et al. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol, 2012, 47(6): 738–745. DOI: 10.1165/rcmb.2012-0088OC |
| [153] | Hunter RC, Asfour F, Dingemans J, et al. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio, 2013, 4(4): e00557–13. |
| [154] | Barton HA, Johnson Z, Cox CD, et al. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol, 1996, 21(5): 1001–1017. DOI: 10.1046/j.1365-2958.1996.381426.x |
| [155] | Matthijs S, Baysse C, Koedam N, et al. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol Microbiol, 2004, 52(2): 371–384. DOI: 10.1111/j.1365-2958.2004.03999.x |
| [156] | Wilderman PJ, Sowa NA, FitzGerald DJ, et al. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA, 2004, 101(26): 9792–9797. DOI: 10.1073/pnas.0403423101 |
| [157] | Vasil ML. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals, 2007, 20(3/4): 587–601. |
| [158] | Cornelis P. Iron uptake and metabolism in Pseudomonas. Appl Microbiol Biotechnol, 2010, 86(6): 1637–1645. DOI: 10.1007/s00253-010-2550-2 |
| [159] | Martin LW, Reid DW, Sharples KJ, et al. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals, 2011, 24(6): 1059–1067. DOI: 10.1007/s10534-011-9464-z |
| [160] | De Vos D, De Chial M, Cochez C, et al. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type Ⅱ pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol, 2001, 175(5): 384–388. DOI: 10.1007/s002030100278 |
| [161] | Lamont IL, Konings AF, Reid DW. Iron acquisition by Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Biometals, 2009, 22(1): 53–60. DOI: 10.1007/s10534-008-9197-9 |
| [162] | Konings AF, Martin LW, Sharples KJ, et al. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun, 2013, 81(8): 2697–2704. DOI: 10.1128/IAI.00418-13 |
| [163] | Singh PK, Parsek MR, Greenberg EP, et al. A component of innate immunity prevents bacterial biofilm development. Nature, 2002, 417(6888): 552–555. DOI: 10.1038/417552a |
| [164] | Rogan MP, Taggart CC, Greene CM, et al. Loss of microbicidal activity and increased formation of biofilm due to decreased lactoferrin activity in patients with cystic fibrosis. J Infect Dis, 2004, 190(7): 1245–1253. DOI: 10.1086/jid.2004.190.issue-7 |
| [165] | Berlutti F, Morea C, Battistoni A, et al. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia. Int J Immunopathol Pharmacol, 2005, 18(4): 661–670. DOI: 10.1177/039463200501800407 |
| [166] | Cornelis P, Wei Q, Andrews SC, et al. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics, 2011, 3(6): 540–549. DOI: 10.1039/c1mt00022e |
| [167] | Wiens JR, Vasil AI, Schurr MJ, et al. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio, 2014, 5(1): e01010–013. |
| [168] | Yu S, Wei Q, Zhao TH, et al. A survival strategy for Pseudomonas aeruginosa that uses exopolysaccharides to sequester and store iron to stimulate Psl-dependent biofilm formation. Appl Environ Microbiol, 2016, 82(21): 6403–6413. |
 2017, Vol. 33
2017, Vol. 33