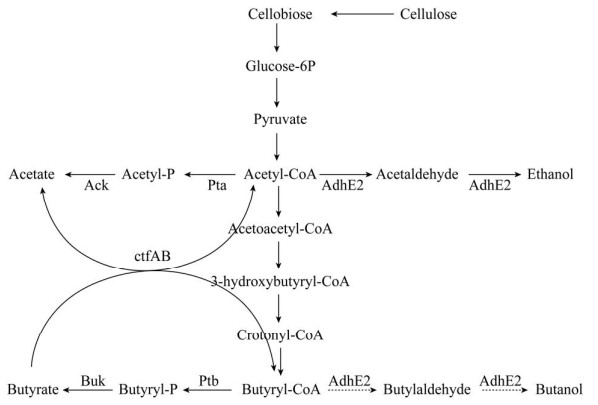
| [1] | |
|
| [2] | |
|
| [3] |
Shanmugam S, Sun CR, Zeng XM, et al. High-efficient production of biobutanol by a novel Clostridium sp. strain WST with uncontrolled pH strategy. Bioresour Technol, 2018, 256: 543-547. DOI:10.1016/j.biortech.2018.02.077 |
|
| [4] | |
|
| [5] |
Sun CR, Zhang SF, Xin FX, et al. Genomic comparison of Clostridium species with the potential of utilizing red algal biomass for biobutanol production. Biotechnol Biofuels, 2018, 11: 42. DOI:10.1186/s13068-018-1044-9 |
|
| [6] |
Zhou ZY, Li Z, Zhang QH, et al. Research progress in production of butanol from lignocellulose. Microbiol China, 2019, 46(12): 3461-3468 (in Chinese). 周智友, 李志, 张庆华, 等. 利用木质纤维素生产丁醇的研究进展. 微生物学通报, 2019, 46(12): 3461-3468. |
|
| [7] |
Jiang Y, Liu JL, Jiang WH, et al. Current status and prospects of industrial bio-production of n-butanol in China. Biotechnol Adv, 2015, 33(7): 1493-1501. DOI:10.1016/j.biotechadv.2014.10.007 |
|
| [8] |
Xiao M, Wu YD, Xue C. Biorefinery and research progress of butanol production. Chin J Bioprocess Eng, 2019, 17(1): 60-71 (in Chinese). 肖敏, 吴又多, 薛闯. 丁醇的生物炼制及研究进展. 生物加工过程, 2019, 17(1): 60-71. |
|
| [9] |
Shukor H, Abdeshahian P, Al-Shorgani NKN, et al. Saccharification of polysaccharide content of palm kernel cake using enzymatic catalysis for production of biobutanol in acetone-butanol-ethanol fermentation. Bioresour Technol, 2016, 202: 206-213. DOI:10.1016/j.biortech.2015.11.078 |
|
| [10] |
Zhang J, Zong WM, Hong W, et al. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab Eng, 2018, 47: 49-59. DOI:10.1016/j.ymben.2018.03.007 |
|
| [11] | |
|
| [12] |
Hijosa-Valsero M, Paniagua-García AI, Díez-Antolínez R. Biobutanol production from apple pomace: the importance of pretreatment methods on the fermentability of lignocellulosic agro-food wastes. Appl Microbiol Biotechnol, 2017, 101(21): 8041-8052. DOI:10.1007/s00253-017-8522-z |
|
| [13] |
Hou XR, From N, Angelidaki I, et al. Butanol fermentation of the brown seaweed Laminaria digitata by Clostridium beijerinckii DSM-6422. Bioresour Technol, 2017, 238: 16-21. DOI:10.1016/j.biortech.2017.04.035 |
|
| [14] |
Xin FX, Wu YR, He JZ. Simultaneous fermentation of glucose and xylose to butanol by Clostridium sp. strain BOH3. Appl Environ Microbiol, 2014, 80(15): 4771-4778. DOI:10.1128/AEM.00337-14 |
|
| [15] | |
|
| [16] |
Qin AD, Duns GJ, Pan T, et al. Consolidated processing of biobutanol production from food wastes by solventogenic Clostridium sp. strain HN 4. Bioresour Technol, 2018, 264: 148-153. DOI:10.1016/j.biortech.2018.05.076 |
|
| [17] |
Xin FX, Chen TP, Jiang YJ, et al. Strategies for improved isopropanol-butanol production by a Clostridium strain from glucose and hemicellulose through consolidated bioprocessing. Biotechnol Biofuels, 2017, 10(1): 118. DOI:10.1186/s13068-017-0805-1 |
|
| [18] |
Yamada R, Hasunuma T, Kondo A. Endowing non-cellulolytic microorganisms with cellulolytic activity aiming for consolidated bioprocessing. Biotechnol Adv, 2013, 31(6): 754-763. DOI:10.1016/j.biotechadv.2013.02.007 |
|
| [19] |
Wen ZQ. Mixed-culture and metabolic engineering of anaerobic clostridia for one-pot butanol production from Alkali extracted corn cobs[D]. Hangzhou: Zhejiang University, 2014 (in Chinese).
闻志强.以碱处理玉米棒芯为原料直接生产丁醇的厌氧梭菌混菌发酵过程及代谢工程研究[D].杭州: 浙江大学, 2014.
|
|
| [20] |
Higashide W, Li YC, Yang YF, et al. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl Environ Microbiol, 2011, 77(8): 2727-2733. DOI:10.1128/AEM.02454-10 |
|
| [21] |
Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature, 2008, 451(7174): 86-89. DOI:10.1038/nature06450 |
|
| [22] |
Gaida SM, Liedtke A, Jentges AHW, et al. Metabolic engineering of Clostridium cellulolyticum for the production of n-butanol from crystalline cellulose. Microb Cell Fact, 2016, 15(1): 6. |
|
| [23] |
Tardif C, Maamar H, Balfin M, et al. Electrotransformation studies in Clostridium cellulolyticum. J Indl Microbiol Biot, 2001, 27(5): 271-274. DOI:10.1038/sj.jim.7000081 |
|
| [24] |
Jang YS, Im JA, Choi SY, et al. Metabolic engineering of Clostridium acetobutylicum for butyric acid production with high butyric acid selectivity. Metab Eng, 2014, 23: 165-174. DOI:10.1016/j.ymben.2014.03.004 |
|
| [25] | |
|
| [26] | |
|
| [27] | |
|
| [28] |
Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng, 2010, 12(4): 307-331. |
|
| [29] | |
|
| [30] |
Jang YS, Lee JY, Lee J, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio, 2012, 3(5): e00314-12. |
|
| [31] | |
|
| [32] |
Biswas R, Zheng TY, Olson DG, et al. Elimination of hydrogenase active site assembly blocks H 2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels, 2015, 8(1): 20. |
|
| [33] |
Yang XR, Xu MM, Yang ST. Metabolic and process engineering of Clostridium cellulovorans for biofuel production from cellulose. Metab Eng, 2015, 32: 39-48. |
|
| [34] |
Jiang YJ, Guo D, Lu JS, et al. Consolidated bioprocessing of butanol production from xylan by a thermophilic and butanologenic Thermoanaerobacterium sp. M5. Biotechnol Biofuels, 2018, 11(1): 89. |
|
| [35] |
Biswas R, Prabhu S, Lynd LR, et al. Increase in ethanol yield via elimination of lactate production in an ethanol-tolerant mutant of Clostridium thermocellum. PLoS ONE, 2014, 9(2): e86389. |
|
| [36] |
Papanek B, Biswas R, Rydzak T, et al. Elimination of metabolic pathways to all traditional fermentation products increases ethanol yields in Clostridium thermocellum. Metab Eng, 2015, 32: 49-54. |
|
| [37] | |
|
| [38] | |
|
| [39] |
Lin PP, Mi L, Morioka AH, et al. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab Eng, 2015, 31: 44-52. |
|
| [40] |
Kylilis N, Tuza ZA, Stan GB, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia. Nat Commun, 2018, 9(1): 2677. |
|
| [41] | |
|
| [42] |
Yu EKC, Chan MKH, Saddler JN. Butanol production from cellulosic substrates by sequential co-culture of Clostridium thermocellum and C. acetobutylicum. Biotechnol Lett, 1985, 7(7): 508-514. |
|
| [43] |
Zhao Q, Characterization and analysis of interaction and biohydrogen between Clostridium thermocellum and Thermoanaerobacterium thermosaccharolyticum in a co-culture system for lignocelluloses degradation[D]. Ji'nan: Shandong University, 2017 (in Chinese).
赵奇.在木质纤维素降解过程中热纤梭菌和热厌氧热解糖杆菌共生关系以及产氢机理的解析[D].济南: 山东大学, 2017.
|
|
| [44] |
Nakayama S, Kiyoshi K, Kadokura T, et al. Butanol production from crystalline cellulose by cocultured Clostridium thermocellum and Clostridium saccharoperbutylacetonicum N1-4. Appl Environ Microbiol, 2011, 77(18): 6470-6475. |
|
| [45] |
Lin YJ, Wen ZQ, Zhu L, et al. Butanol production from corncob in the sequential co-culture of Clostridium thermocellum and Clostridium beijerinckii. J Chem Eng Chin Univ, 2013, 27(3): 444-449 (in Chinese). 林逸君, 闻志强, 朱力, 等. Clostridium thermocellum与 Clostridium beijerinckii偶联发酵玉米棒芯产丁醇. 高校化学工程学报, 2013, 27(3): 444-449. |
|
| [46] |
Nakayama S, Bando Y, Ohnishi A, et al. Decreased hydrogen production leads to selective butanol production in co-cultures of Clostridium thermocellum and Clostridium saccharoperbutylacetonicum strain N1-4. J Biosci Bioeng, 2013, 115(2): 173-175. |
|
| [47] | |
|
| [48] | |
|
| [49] | |
|
| [50] |
Fu DF. Isolation of butanol production strain and construction of co-culture system with cellulose[D]. Harbin: Harbin Institute of Technology, 2016 (in Chinese).
傅德丰.丁醇生产菌的筛选及以纤维素为底物共培养体系的构建[D].哈尔滨: 哈尔滨工业大学, 2016.
|
|
| [51] | |
|
| [52] |
Wu JJ, Du GC, Zhou JW, et al. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng, 2013, 16: 48-55. |
|
| [53] |
Wen ZQ, Minton NP, Zhang Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab Eng, 2017, 39: 38-48. |
|
| [54] | |
|
| [55] | |
|
| [56] | |
|
| [57] |
Qian YC, Zhong LX, Gao J, et al. Production of highly efficient cellulase mixtures by genetically exploiting the potentials of Trichoderma reesei endogenous cellulases for hydrolysis of corncob residues. Microb Cell Fact, 2017, 16(1): 207. |
|
| [58] |
Derntl C, Guzmán-Chávez F, Mello-de-Sousa TM, et al. In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei. Front Microbiol, 2017, 8: 2037. |
|
| [59] | |
|
| [60] | |
|
 2020, Vol. 36
2020, Vol. 36