中国科学院微生物研究所、中国微生物学会主办
文章信息
- 韩群, 秦亚玲, 李德峰
- Han Qun, Qin Yaling, Li Defeng
- 细菌Rieske非血红素铁环羟化酶在多环芳烃降解中的研究进展
- Advances in bacterial Rieske non-heme iron ring-hydroxylating dioxygenases that initiate polycyclic aromatic hydrocarbons degradation
- 生物工程学报, 2021, 37(10): 3439-3458
- Chinese Journal of Biotechnology, 2021, 37(10): 3439-3458
- 10.13345/j.cjb.210402
-
文章历史
- Received: May 31, 2021
- Accepted: September 1, 2021
- Published: September 1, 2021
2. 中国科学院大学生命科学学院,北京 100049
2. College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China
多环芳烃(Polycyclic aromatic hydrocarbons,PAHs) 是一种常见的持久性有机污染物,其分子结构中含有两个及以上苯环,疏水性强,水溶性低,能够稳定地存在于环境中,不易被降解[1],因其具有致癌、致突变等毒性而被广泛关注[2]。多环芳烃根据分子量的高低,分为由萘、菲、蒽等2个或3个稠环组成的低分子量多环芳烃和由荧蒽、芘、苯并[a]蒽和苯并[a]芘等4个或4个以上环组成的高分子量多环芳烃[3]。在国内外,多环芳烃都是非常受关注的环境污染物。1995年,美国环保署就将16种多环芳烃列为优先控制的污染物[4],我国《土壤环境质量建设用地土壤污染风险管控标准(试行)》 (GB36600-2018)[5]等国家标准将萘、苯并[a]蒽等8种PAHs列入环境污染风险筛选和管控范围(图 1)。目前,多环芳烃有物理、化学和生物等降解方式[6]。其中,生物降解是一种自然降解方式,占环境中多环芳烃降解的40%–60%[7],是去除或者降低环境中多环芳烃污染的一种重要方式[8]。当前已经从多环芳烃污染的环境中分离到多种以多环芳烃为唯一碳源的菌株,如能降解萘、菲的Diaphorobacter sp.、寡养单胞菌属Stenotrophomonas sp.[9-10],以及能够降解高分子量多环芳烃荧蒽、芘的红球菌属Rhodococcus sp.、假单胞菌属Pseudomonas sp.、鞘氨醇盒菌属Sphingopyxis sp.、芽孢杆菌属Bacillus sp.[1, 11]等。国内很多实验室也分离获得对萘[12]、菲、荧蒽[13]、苯并[a]芘[14]等多种多环芳烃具有高效降解作用的菌株,比如恶臭假单胞菌Pseudomonas putida B6-2对13种多环芳烃及7种二噁英都具有一定的降解能力[15],而油菜假单胞菌Pseudomonas brassicacearum MPDS对多环芳烃和杂环衍生物都具有高效的降解能力[16]。另外,我国科学家在多环芳烃污染的来源、时间空间分布[17]、健康风险评估[18-19]、生物修复[20]、代谢途径(如下游水杨酸途径[21]、龙胆酸代谢途径[22]等) 以及相关酶(如环羟基化酶[23]等) 分子机制研究等方面做了大量的突出工作。目前低分子量多环芳烃及其下游代谢途径的研究已经比较清楚,尤其是萘、菲、蒽、芴等的降解途径[24]。而由于高分子量多环芳烃的水溶性更差、代谢途径更长,也更复杂多样化[25],对于细菌中高分子量多环芳烃降解途径的研究不甚透彻,在已经报道的高分子量多环芳烃降解菌株中的具体代谢途径及其中所涉及的酶的鉴定及其性质等还需进一步探究。可以预见在较长一段时间内,针对高致畸性高危害性高分子量多环芳烃的降解机制和代谢途径研究将是多环芳烃研究的重点。
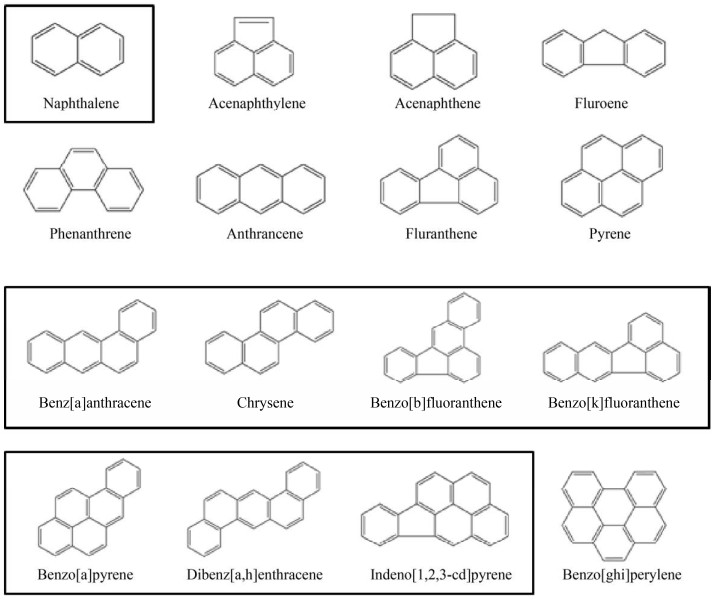
|
| 图 1 美国环保署优先控制的16种多环芳烃 Fig. 1 Sixteen PAHs regulated by EPA of US. (黑色框为国内管控的8种多环芳烃) (black boxed are the eight PAHs regulated in China) |
| |
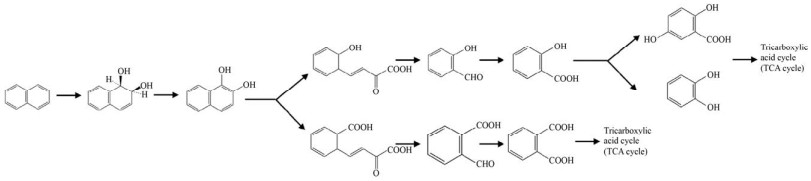
多环芳烃的降解起始于羟化,然后脱氢、开环、一步步去除支链,最终进入三羧酸循环(Tricarboxylic acid cycle,TCA cycle) 被微生物所利用。微生物降解多环芳烃的第一步反应即羟化反应,这一步在细菌中研究较多的酶是Rieske非血红素铁环羟基化酶(Rieske-type non-heme iron aromatic ring-hydroxylating dioxygenases,RHOs),在真菌中研究较多的是细胞色素P450单加氧酶(Cytochrome P450 monooxygenase,CYP450)。RHOs能催化多种氧化反应,包括碳-碳键的氧化裂解、单羟基化和二羟基化反应[26]。这种酶是唯一已知的催化邻位顺式二醇立体选择性一步形成的酶[27],能在一个酶促步骤中将两个羟基引入到多环芳烃上形成顺式二氢二醇(图 2)[28]。顺式二氢二醇经过脱氢酶脱氢后形成相应的二醇,在内二醇或外二醇双加氧酶的作用下开环,内二醇双加氧酶作用于两个羟基之间的C-C键,而外二醇双加氧酶切割与两个羟基相邻的C-C键底物[29]。例如,PhdF就是一种Ⅰ型外二醇双加氧酶,是邻位氧螯合超家族的成员,可作用于3, 4-二羟基菲等含有双羟基的苯环上,发挥开环的功能[30]。CYP450在人类、植物、微生物中是广泛存在的,而且其底物范围非常广泛,能够催化C-羟基化及杂原子氧合、释放和环氧化物的形成等[31]。CYP450参与了PAHs生物降解过程的初始氧化步骤,催化多环芳烃的不同位置形成一种或多种单羟基化产物[32],或者与环氧化物水解酶协同作用催化形成反式-二氢二醇(图 3)[33-34]。CYP450主要在真菌中研究的较多,如白腐真菌[35]、黄孢原毛平革菌,且能够作用于四环、五环乃至六环的苯并[ghi]苝等高分子量的多环芳烃[36],细菌中少见报道。而在Rhodococcus sp. P14[37]和Mycobacterium vanbaalenii PYR-1[38]中都提到存在RHOs与CYP450共同降解多环芳烃的现象,这些菌能同时催化高分子量PAHs产生顺式-二氢二醇和反式-二氢二醇。而且研究者证明在多株Mycobacterium vanbaalenii菌中都有RHOs与CYP450基因共同存在的情况[38]。前面提到,内二醇以及外二醇双加氧酶只能识别两个羟基中间或者与两个羟基相邻的C-C键,而CYP450作用多环芳烃后一部分仅产生一个羟基形成酚,因而推测在CYP450羟化后,可能由CYP450再进行一次羟化,或者由RHOs对CYP450单羟基化的产物再次羟基化,最终形成开环酶能够作用的二醇的结构。而且早期也有文章提到RHOs能够催化单加氧反应[39],如甲苯单加氧酶可能催化苯酚生成邻苯二酚[40-41],萘双加氧酶也被证实能参与多种苄基单羟基化反应[42]。也有证据表明萘双加氧酶能够催化荧蒽和芴单羟基化分布形成8-羟基荧蒽和9-芴醇[43],以及其他的研究中RHOs催化的单羟基化产物的出现[44]。RHOs作为高分子量多环芳烃降解的两类起始酶之一,一旦确定特定多环芳烃可以被某一RHOs氧化后,即有可能根据基因组分布、代谢物分析等推测相关代谢中间物和中间途径,具有“提纲挈领”的意义。但是RHOs的生化研究过程中存在诸多需要被克服的困难。从技术上来说,首先,多环芳烃特别是高分子量多环芳烃溶解性差[24],在体外酶活系统构建中与酶的结合能力差,需要有机溶剂溶解,但有机溶剂又可能对酶活性造成损伤[43]。RHOs体外酶活系统中所需的电子传递体中的铁氧还蛋白还原酶(Ferredoxin reductase,FdR) 难以纯化表达[43],给体外酶活测定带来一定的困难,选择外源的电子传递体不能保证双加氧酶的酶活效率,在确定合适的电子传递体前,酶的活性难以用传统的NADH的消耗来表征;另外,由于高分子量的多环芳烃环数多,RHOs作用的位置难以确定,产生一种甚至多种产物[45],而对于产物的鉴定多采用的是气相色谱-质谱联用技术(GC-MS),因为产物缺少标准品或者标准品价格昂贵,使得研究方法受限。而在生信分析中,RHOs的底物范围非常广泛,以萘双加氧酶(Naphthalene dioxygenase,NDO) 为例[42],它可以以多种物质为底物,这也为酶的底物确定带来了一定的困难。
|
|
图 2 RHOs起始催化萘的降解途径
Fig. 2 Degradation pathway of naphthalene initiated by RHOs.
|
| |

|
|
图 3 CYP450催化菲
Fig. 3 Catalytic activity of CYP450 on phenanthrene.
|
| |
综上,细菌RHOs在多环芳烃的降解途径中发挥着非常重要的作用,也存在着一定的研究难度和迫切之处,下面我们将对与多环芳烃降解相关的RHOs进行详细的论述。
2 Rieske非血红素铁环羟基化双加氧酶(RHOs) 的来源、组成 2.1 RHOs的菌株来源分布目前对于RHOs的研究集中在Rhodococcus sp.、Pseudomonas sp.、罗尔斯通氏菌属Ralstonia sp.、海杆菌属Marinobacter sp.、丛毛单胞菌属Comamonas sp.、潘多拉菌属Pandoraea sp.等菌株中[46]。表 1统计了1988年至今能够使用如Naphthalene dioxygenase、Biphenyl dioxygenases等特定关键词查询到的在文献中发表的已经与多环芳烃降解相关的RHOs基因,少有发现新型的RHOs,进行表征的都与早期发现的RHOs具有高度同源性。在多环芳烃降解菌的筛选及研究过程中还发现RHOs可能在不同菌属中存在水平转移的现象。早期有观点认为RHOs可能起源于革兰氏阴性细菌,随后转移到革兰氏阳性细菌中[47]。水平转移的现象被研究者们多次提到,如:利用简并PCR引物从Marinobacter中扩增萘双加氧酶的大亚基,经系统发育树分析,该萘双加氧酶与Pseudomonas和伯克氏菌属Burkholderia来源的萘双加氧酶相似,而在其基因簇附近发现了tnpA1基因的同源基因,可能编码噬菌体λ型转座酶,这可能解释了多环芳烃降解基因在这些细菌谱系中发生的水平转移[48]。同样的,以来自Pseudomonas spp.的2Fe-2S还原酶基因设计引物,发现食酸菌属Acidovorax中有与Pseudomonas spp.相同的萘双加氧酶的相关基因,但是属于同一物种的菌株则没有[49]。研究推测这种基因水平转移的发生在微生物群落适应污染物中发挥了重要作用[50]。
| GenBank | Strains | RHOs | Substrates | Year | References |
| CP054128 | Pseudomonas sp. MPDS | Naphthalene dioxygenase | Naphthalene, fluorene, dibenzofuran and dibenzothiophene | 2021 | [16] |
| MH560349 | Pseudomonas fluorescens AH-40 | Naphthalene dioxygenase | Phenanthrene | 2020 | [51] |
| KJ461700 | Pseudomonas aeruginosa N6P6 | Naphthalene dioxygenase | Naphthalene | 2020 | [52] |
| MULN00000000 | Pseudomonas veronii strainVI4T1 |
Naphthalene dioxygenase | Naphthalene | 2019 | [53] |
| JN613334 | Rhodococcus wratislaviensis strain 9 | Naphthalene dioxygenase | Phenanthrene | 2019 | [54] |
| AAD28100 | Rhodococcus sp. strain NCIMB12038 | Naphthalene dioxygenase | Naphthalene | 2019 | [55] |
| CP006254 | Geobacillus sp. JF8 | Naphthalene dioxygenase | Naphthalene | 2019 | [56] |
| NO | Sphingobium yanoikuyae B1 | Biphenyl 2, 3-dioxygenase | Phenazine | 2017 | [57] |
| KC771235 | Pseudomonas aeruginosa JP-11 | Biphenyl dioxygenase | Biphenyl | 2016 | [58] |
| JN235141 | Rhodococcus sp. ustb-1 | Naphthalene dioxygenase | Pyrene | 2015 | [59] |
| Q46372 | Pandoraea pnomenusa B356 | Biphenyl dioxygenases | 3-hydroxy-4, 4′-dichlorobiphenyl, 3, 3′-dihydroxy-4, 4′-chlorobiphenyl, flavone, isoflavone, and flavanone | 2015 | [60] |
| KM102520 |
Uncultured gammaproteobacterium | Aromatic ring-hydroxylating oxygenases (phd20/19) |
Biphenyl, naphthalene, phenanthrene, pyrene, fluoranthene | 2014 | [61] |
| KM102522 |
Uncultured gammaproteobacterium | Aromatic ring-hydroxylating oxygenases (bph29/28) |
Biphenyl, naphthalene | 2014 | [61] |
| KM102523 |
Uncultured gammaproteobacterium | Aromatic ring-hydroxylating oxygenases (nah33/32) |
Biphenyl, naphthalene, phenanthrene | 2014 | [61] |
| JN655512 | Comamonas sp. MQ | Naphthalene dioxygenase | Indole and most indole derivatives | 2013 | [62] |
| NO | Martelella sp. AD-3 | Naphthalene dioxygenase | Anthracene | 2012 | [63] |
| P37333 | Pseudomonas strain LB400 | A variant biphenyl dioxygenase | Dibenzofuran | 2012 | [64] |
| GQ184726 | Burkholderia sp. C3 | Nag-like dioxygenases | Naphthalene, dibenzothiophene | 2011 | [65] |
| GQ184727 | Burkholderia sp. C3 | Phn-like dioxygenases | Naphthalene, phenanthrene, dibenzothiophene | 2011 | [65] |
| Q53122 | Rhodococcus jostii RHA1 | Biphenyl and ethylbenzene dioxygenases | Polybrominated diphenyl ethers | 2011 | [66] |
| SRA028415 | Sediment metagenome | Novel aromatic ring hydroxylating dioxygenases | Biphenyls | 2011 | [67] |
| DQ846881 | Rhodococcus opacus R7 | Naphthalene dioxygenase | Naphthalene | 2010 | [68] |
| Q3C1D5 | Comamonas sp. strain E6 | Terephthalate 1, 2-dioxygenase | Terephthalate | 2008 | [69] |
| Q53122 | Rhodococcus jostii RHA1 | Biphenyl and ethylbenzene dioxygenases | Styrene and benzene | 2008 | [70] |
| Q53122 | Rhodococcus jostii RHA1 | Biphenyl 2, 3-dioxygenase | Biphenyl/polychlorinated- biphenyl | 2007 | [71] |
| HE577117 | Uncultured bacterium | Biphenyl dioxygenases | Biphenyls | 2007 | [72] |
| EF152282 | Sphingobium yanoikuyae B1 | Biphenyl/naphthalene dioxygenase | Biphenyl, naphthalene, and phenanthrene, toluene, m- and p-xylene | 2007 | [73] |
| AF295032 | Marinobacter strain NCE312 | Naphthalene dioxygenase | Naphthalene and 2-methylnaphthalene | 2006 | [48] |
| Q46372 | Pandoraea pnomenusa B356 | Biphenyl dioxygenases | 2-hydroxy-3-chlorobiphenyl, 2-hydroxy-5-chlorobiphenyl and 2-hydroxy-3, 5-dichlorobiphenyl | 2004 | [74] |
| P37333 | Pseudomonas strain LB400 | Biphenyl dioxygenases | 2-hydroxy-3-chlorobiphenyl, 2-hydroxy-5-chlorobiphenyl and 2-hydroxy-3, 5-dichlorobiphenyl, 2, 2′-dichlorobiphenyl | 2004 | [74-75] |
| O52382 | Ralstonia sp. strain U2 | Naphthalene dioxygenase | Naphthalene | 2002 | [76] |
| AF061751 | Burkholderia sp. strain RP007 | Phn-like dioxygenases | Naphthalene, phenanthrene | 1999 | [77] |
| Q46372 | Comamonas testosteroni strain B-356 | Biphenyl dioxygenases | Biphenyl/chlorobiphenyl dioxygenase | 1996 | [78] |
| A5W4F2 | Pseudomonas putida F1 | Toluene dioxygenase | Benzene and toluene | 1994 | [79] |
| Q52438 | Pseudomonas sp. strain KKS102 | Biphenyl 2, 3-dioxygenase | Biphenyl and polychlorinated biphenyls | 1994 | [55] |
| Q07944 | Pseudomonas putida ML2 | Benzene dioxygenase | Benzene | 1993 | [80] |
| Q52028 | Pseudomonas pseudoalcaligenes KF707 | Biphenyl dioxygenase | Biphenyls and polychlorinated biphenyls | 1992 | [81] |
| P37333 | Pseudomonas strain LB400 | Biphenyl dioxygenases | Polychlorinated-biphenyl | 1992 | [82] |
| P0A110 | Pseudomonas putida strain NCIB9816 | Naphthalene dioxygenase | Indole | 1988 | [83] |
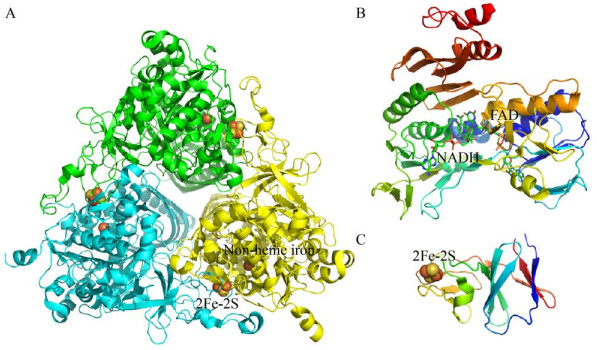
Rieske非血红素铁环羟基化酶(RHOs) 是一种多组分酶系统,由末端双加氧酶、铁氧还蛋白(Ferredoxin,Fd)、铁氧还蛋白还原酶(Ferredoxin reductases,FdR)[84]三部分组成(图 4)。末端加氧酶一般含有大亚基和小亚基,构成α3β3异六聚体结构,如Sphingobium yanoikuyae B1中的联苯双加氧酶[85];或者是只含有大亚基,构成α3同三聚体,如咔唑的末端加氧酶成分(PDB ID:4NBD)[86]。末端双加氧酶的α亚基包含一个Rieske [2Fe-2S]簇和一个单核铁活性中心。Fd含有一个Rieske型的[2Fe-2S]簇[87],而FdR是由3个结构域组成:FAD结合结构域、NADH结合结构域和C末端结构域[88],有些菌中的Fd和FdR可能融合形成一个蛋白,如食醚红球菌Rhodococcus aetherivorans IcdP1[89] (GenBank登录号:CP011341) 中的RHOs加氧酶组分附近分布着一些潜在的天然融合NAD/FAD或FMN/NAD结合域和[2Fe-2S]簇的蛋白,可能是为RHOs的加氧酶组分发挥电子传递作用,这种融合蛋白发挥电子传递作用早在1992年就有报道,如黄素蛋白与[2Fe-2S]簇的天然融合[90]。RHOs的3个组分行使功能时,首先是FdR从NAD(P)H中释放电子,并将电子转移到Fd中,Fd将电子转移到加氧酶[91],加氧酶的Rieske [2Fe-2S]簇从电子转移组分接受电子后,将电子转移到活性位点的单核铁,激活分子氧,从而攻击底物[26]。
|
| 图 4 RHOs的各组成部分的代表性结构 Fig. 4 Structures of three RHOs components. (A) Dioxygenase component (PDB ID: 2CKF). (B) Ferredoxin reductase (PDB ID: 1F3P). (C) Ferredoxin (PDB ID: 2QPZ). |
| |
萘双加氧酶(Naphthalene dioxygenase,NDO)是与多环芳烃降解相关的Rieske非血红素铁环羟化酶中研究较多的一类。萘双加氧酶有多种类型,nag类基因是从Pseudomonas putida strain G7首次发现的能够降解萘的基因,phn是从Burkholderia sp. RP007中发现的能以萘、菲和蒽为唯一碳源的萘降解基因[65]。NDO的底物范围非常广,可作用于萘之外的多种物质,具体可参考表 1所统计的部分RHOs的底物。例如,荧光假单胞菌Pseudomonas fluorescens AH-4、Rhodococcus wratislaviensis strain 9的NDO能氧化菲[51, 54]。萘双加氧酶还能作用于吲哚产生靛蓝或靛玉红[92];鞘氨醇单胞菌Sphingomonas CHY-1中的NDO能羟化荧蒽、苯并[a]蒽、䓛、苯并[a]芘、芘,其中,苯并[a]芘的分子量虽然比芘更大,但是NDO作用于苯并[a]芘时的酶活更高,可能是在与NDO接触时,苯并[a]芘多出来的一个环更能靠近狭窄的酶活性中心[93]。另外RHOs中还有一类与联苯型多环芳烃相关的联苯双加氧酶(Biphenyl dioxygenase,BPDOs) 的研究也相对较多。BPDOs作用的底物多与多氯联苯及其衍生物相关,如Pandoraea pnomenusa B356的BPDOs能够催化联苯,后续发现它可将5-氯-2-羟基联苯转化为5-氯-2-羟基苯甲酸酯,还可参与对双对位氯取代联苯类似物(3-羟基-4, 4′-二氯联苯和3, 3′-二羟基-4, 4′-氯联苯) 的代谢[60]。Pseudomonas strain LB400也可降解联苯,同B-356 BPDOs一样具有可以催化2-羟基-3-氯联苯、2-羟基-5-氯联苯和2-羟基-3, 5-二氯联苯的能力[74]。另外LB400还具有催化2, 2′-二氯联苯的能力[75]。除此之外,有研究者还发现Rhodococcus jostii RHA1中的联苯和乙苯双加氧酶在联苯、乙苯等底物存在下能够转化多溴二苯醚(PBDEs)[66]。
4 基于结构生物学的底物识别机制研究RHOs可以催化多种反应,与许多不同底物形成复合物的结构表明,活性位点中底物的取向不仅控制区域特异性和立体特异性,而且还控制催化的反应类型[94]。在RHOs结合底物后,没有发现明显的侧链重排[91],底物与单核铁活性中心接近的两个碳被羟基化,这解释了由RHOs产生的顺式二氢二醇的极端区域和立体选择性[91]。底物必须与RHOs充分相互作用,以防止催化时底物发生运动,催化活性口袋内部若存在多余空间可能会影响RHOs在发生催化作用时底物在口袋内部的活动[95]。
表 2所示是PDB数据库中部分已解析结构的RHOs。RHOs利用一个单核非血红素铁中心来进行催化,此过程消耗了2个电子、2个质子和1个氧气分子,产生了顺式二氢二醇。NDO的结构表明活性位点口袋内的氨基酸大部分是疏水性的,这为芳香族底物的结合提供了合适的环境[96]。Pseudomonas sp. NCIB 9816-4的NDO结构是第一个被解析的RHOs[97]。它的第352位突变影响了萘的立体选择性,206位和295位影响了联苯和菲的区域选择性,在同时突变206/352或者206/295/352的情况下,NDO催化菲形成菲顺式-9, 10-二氢二醇作为主要产物[98],表明催化位点附近的关键残基影响RHOs的底物识别。
| PDB ID | Strains | RHOs | Substrates | Year | References |
| 4HJL,
4HKV, 4HM0-8 |
Pseudomonas sp. C18 | Naphthalene 1, 2-dioxygenase | 1-chloronaphthalene, benzamide, indole-3-acetate, thioanisole, styrene, indene, phenetole, indan, ethylbenzene, ethylphenylsulfide, 1-indanone | 2012 | [103] |
| 2YFI | ParaBurkholderia xenovorans LB400 | Biphenyl dioxygenase variant Rr41 | Dibenzofuran | 2011 | [104] |
| 2XR8 | ParaBurkholderia xenovorans LB400 | Biphenyl dioxygenase | Many polychlorinated biphenyls | 2010 | [105] |
| 3EN1 | Pseudomonas putida | Toluene 2, 3-dioxygenase | Toluene | 2010 | [88] |
| 3GZY | Comamonas testosteroni sp. strain B-356 | Biphenyl dioxygenase | Biphenyl and polychlorinated biphenyls | 2009 | [106] |
| 2CKF | Sphingomonas sp. CHY-1 | PAH-hydroxylating dioxygenase | Fluoranthene, benz[a]anthracene, benzo[a]pyrene | 2007 | [99] |
| 2HMM | Pseudomonas sp. | Naphthalene 1, 2-dioxygenase | Anthracene | 2006 | [91] |
| 2HML | Pseudomonas sp. | Naphthalene 1, 2-dioxygenase mutant | Phenanthrene | 2006 | [91] |
| 2HMK | Pseudomonas sp. | Naphthalene 1, 2-dioxygenase | Phenanthrene | 2006 | [91] |
| 2GBX | Sphingomonas yanoikuyae B1 | Biphenyl 2, 3-dioxygenase | Biphenyl | 2006 | [100] |
| 2DE7 | Janthinobacterium | Carbazole 1, 9a-dioxygenase | Carbazole | 2006 | [107] |
| 2B24 | Rhodococcus sp. NCIMB 12038 | Naphthalene 1, 2-dioxygenase | Indole | 2005 | [108] |
| 2BMR | Comamonas sp. JS765 | Nitrobenzene dioxygenase | 3-nitrotoluene | 2005 | [109] |
| 1WQL | Pseudomonas fluorescens Ip01 | Cumene dioxygenase | Cumene or toluene | 2004 | [110] |
| 1WW9 | Janthinobacterium sp. J3 | Carbazole 1, 9a-dioxygenase | Carbazole | 2005 | [111] |
| 1UUV | Pseudomonas putida | Naphthalene 1, 2-dioxygenase | Nitric oxide and indole | 2004 | [112] |
| 1ULI | Rhodococcus jostii RHA1 | Biphenyl dioxygenase | Biphenyl | 2003 | [113] |
| 1O7G | Pseudomonas putida | Naphthalene 1, 2-dioxygenase | Naphthalene | 2002 | [114] |
| 1EG9 | Pseudomonas putida | Naphthalene 1, 2-dioxygenase | Indole | 2000 | [115] |
| 1NDO | Pseudomonas putida | Napthalene 1, 2-dioxygenase | Naphthalene | 1998 | [116] |
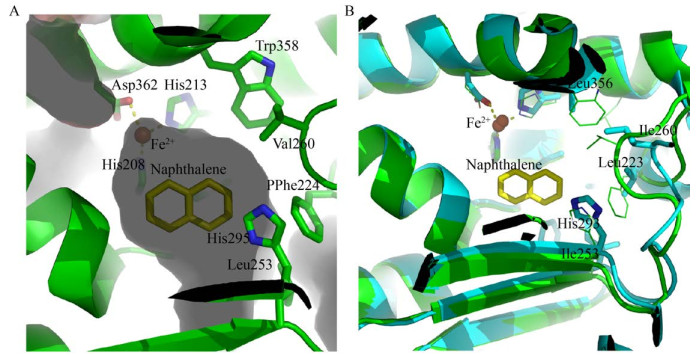
相对于催化萘的RHOs (图 5A) 来说,结合高分子量多环芳烃的RHOs的疏水性底物的结合口袋明显更大,如Sphingomonas CHY-1的RHOs,CHY-1的RHOs是比较典型的能够羟基化五环PAH的酶,它的催化腔的中心区域主要由Phe350、Phe404和Leu356的侧链形成,形成相当均匀的梯形腔,从而影响酶的位置特异性[99]。另外,天然和联苯结合形式的BPDO-OB1结构与萘1, 2-双加氧酶相似,但活性位点入口也明显大于萘1, 2-双加氧酶的入口,活性位点残基的差异也允许高分子量多环芳烃䓛和苯并[a]芘的结合[100]。除此之外,RHOs活性位点入口存在的柔性环也被认为是一种可以扩大底物特异性范围的特征,这个环可以在需要时提供重塑活性位点所需的结构灵活性,有利于不同配体的调节[61]。CHY-1的RHOs的柔性环L1上的Leu223和L2上的Ile260有助于催化位点对高分子量多环芳烃的选择性(图 5B)。同样,在BphAELB400的α亚基上的残基也有一段覆盖活性位点的环,在底物结合时,该片段发生了明显的位移[95]。在鞘氨醇菌属Sphingobium sp. FB3中RHOs的加氧酶组分,223位是Phe,比能羟基化五环PAH的Sphingomonas CHY-1的RHOs的Leu大得多,突变该位点后FB3-RHOs降解苯并[a]芘速率加快[14]。这也说明关键残基的改变就可能改变酶的底物特异性,再如BphAEs从280到283位于催化腔的入口,而283位的Ser被Met取代后,改变了催化位点内底物的方向,从而改变了其羟基化位点,增强了它对于多种多氯联苯的特异性。也正说明283位的突变对BPDO的底物特异性有显著影响[101];BPDO中与底物识别和区域特异性相关的位点被突变后也改变了BPDO在苯环上的作用位点[102]。
|
| 图 5 RHOs底物结合口袋 Fig. 5 The ligand binding pocket of RHOs. (A) The ligand binding pocket of NDO (PDB ID: 1O7G) from Pseudomonas sp. NCIB 9816-4. The naphthalene molecule was shown in yellow and some important residue in green. (B) Comparison with RHOs from Sphingomonas CHY-1 (Blue, PDB ID: 2CKF) and from Pseudomonas sp. NCIB 9816-4 (green). Residues Leu223 and Ile260 with smaller amino acid side chains indicated the possibility of that Sphingomonas CHY-1 RHOs bind PAHs molecules with larger molecular weight. |
| |
用唯一碳源的无机盐培养基从污染环境中富集分离多环芳烃降解菌株是普遍采用的方式。通过污染物压力筛选能产生与多环芳烃降解相关的微群落,进而研究其中的RHOs[117]。环境中还存在着非常多的不可培养微生物发挥降解作用,因此研究人员利用分子生物学的方法来筛选和检测环境中的RHOs,如建立RHOs的加氧酶组分的α-亚基基因序列数据库,以保守的Rieske中心的核酸序列,设计高度特异性的引物以进行RHOs的丰度或多样性等的研究[118]。一系列基于保守区域引物的技术被应用,如聚合酶链式反应(Polymerase chain reaction,PCR)[119]、利用双加氧酶探针进行Southern杂交实验[120]、多重PCR技术等来鉴定和检测芳香族双加氧酶基因,以及利用定量PCR扩增技术量化环境样品中的芳香族分解代谢基因[121]。检测到环境中存在的RHOs后,通过构建宏基因组文库,如以Fosmid、Cosmid、BAC为载体的文库构建,可以获取到相关RHOs的全长序列[122],能够对其功能进行研究及表征,扩展对于未培养微生物中PAH降解基因的了解,更能够扩大对于RHOs的认知,找到更多功能强大的RHOs来用于场地或相关环境的修复。
5.2 大肠杆菌异源表达-体内活性测定RHOs功能表征的一个研究思路是将RHOs的双加氧酶组分、电子传递组分分别克隆到不同质粒上,转到同一宿主中进行诱导表达,若要检测这些基因是否正确表达产生了有降解活性的RHOs,则可以采用靛蓝筛选法[123]、高效液相色谱(HPLC) 或气相色谱(GC) 检测代谢物,使用氧气电极检测实时活性[124]。
其中靛蓝筛选法是最为方便快捷的方法,萘双加氧酶型RHOs的特征是吲哚向靛蓝转化,添加不高于4 mmol/L的吲哚(高于4 mmol/L对细胞有毒性) 能使得菌株中的活性萘双加氧酶(NDO) 生成靛蓝[125],大肠杆菌细胞可以利用色氨酸酶从色氨酸合成吲哚,所以含有RHOs的克隆不需要添加吲哚即可验证在大肠杆菌中的活性[126],从而作为筛选NDO的表征。而HPLC或GC-MS (质谱) 联用是最常用的多环芳烃降解产物的检测和鉴定方法[3, 93, 127]。但该方法操作较为复杂,要对代谢产物经过反复萃取-旋蒸浓缩-干燥除水-重溶-过滤等步骤,不适合高通量的筛选,但是能较为精确地定量。基于高碘酸盐荧光检测RHOs产生的顺式二醇代谢物的方法是一种新的筛选方法,其采用高碘酸钠氧化顺式二醇代谢物转化为相应的二醛,用以产生可检测的分析物[128]。此方法与传统方法相比优势在于能够高通量地筛选双加氧酶或者有羟基化多环芳烃能力的突变体双加氧酶。
5.3 重组蛋白的体外活性与底物结合RHOs的体外酶活测定需要完整的酶组分才能进行,因而需要将加氧酶组分和Fd、FdR电子传递组分都进行纯化表达,得到纯度较高的蛋白质,通过在340 nm处NADH的消耗来测定酶活性[61],或者利用氧电极测定耗氧速率[129]。此外,差示扫描荧光法(Differential scanning fluorimetry,DSF) 是一种基于蛋白质的热稳定性原理高通量筛选蛋白质与配体相互作用的方法,在其他蛋白质的配体筛选中已经被广泛使用。此种方法结合上文提到的高通量的荧光检测方法来检测RHOs与多环芳烃的混合物孵育后的产物中是否有顺式二醇代谢物,不失为一种简单的检测方法,为RHOs的底物筛选提供了一种高通量的方式。
而重组蛋白测活的难点在于FdR的表达存在问题,大部分重组蛋白表达为包涵体的形式,而这个问题不能通过改变宿主菌株或在诱导过程中降低温度来解决[43],睾丸酮丛毛单胞菌Comamonas testosterone B-356联苯双加氧酶的FdR组分表达时也遇到了相同的问题,最后使用QIAGEN His-Binding Kit纯化得到能用于实验的蛋白量[130]。从类产碱假单胞菌Pseudomonas pseudoalcaligenes KF707中克隆表达联苯双加氧酶的FdR组分时,用了一个含有分子伴侣groELS基因的质粒pKY206成功表达了可溶性的FdR蛋白[131]。Sphingomonas sp. strain CHY-1的FdR组分克隆到pET15b载体上转化到大肠杆菌BL21(DE3) 中进行体内活性测定时,发现比没有Fd、FdR组分的菌株对菲的降解率增加了35倍,说明FdR在大肠杆菌中正确合成并发挥作用[132],但后期纯化时只能获得少量的该酶,也不能进行大量的表达,因而更换了其他菌株中更为稳定的FdR进行实验,并不影响电子传递作用[43]。以Pseudomonas putida F1中的甲苯双加氧酶进行了实验验证,在更换Fd的情况下,不更换FdR能获得原来活性的56%,而更换合适的FdR也能使得活性达到原来的38%[84]。因而当FdR不能正确表达时,可以使用分子伴侣辅助FdR表达,也可尝试更换其他来源的FdR电子传递组分进行酶活测定。
5.4 分子模拟通过同源建模、分子对接模拟分析多环芳烃与双加氧酶活性中心的相互作用机理从而解释实验现象是一种比较普遍的方法[23]。研究RHOs底物特异性的方法已经不仅仅局限于实验结果,从计算机的分子模拟中可获得底物和酶的结构信息,并可以解释生化实验中的一些现象,为实验提供参考性的帮助,而且通过对接分析预测的大多数底物是可以通过实验验证的[126]。目前的同源建模可以采用Geno3D、SWISS-MODEL、PHYRE、MODELLER和Schrödinger。建模的结果可通过PROCHECK[59]获取到Ramachandran图来评分,或者通过VADAR软件评估单个残基和整个蛋白质的关键结构参数来评估模型的质量,结构错误可通过WHAT-CHECK进行分析[126],氨基酸序列与其3D原子模型之间的相容性通过VERIFY-3D进行评估[59, 129]。MOLE 2.0识别从溶剂区域通向活性部位的可能通道[133],POCASA 1.0可预测催化口袋的区域[126],分子对接的软件可使用AutoDock Vina[134]或者Sybyl7.3中的Surflex模块,用于探索NDO活性位点与配体之间的相互作用机理,Surflex可以有效地减少假阳性结果的百分比[59]。分子模拟在RHOs的研究中是一个非常好的工具,它能够在获得蛋白质与多环芳烃复合物结构之前推测蛋白与底物的相互作用。对RHOs进行设计改造以产生更有效的酶活,适应环境实际需求来进行环境修复方面,分子模拟是非常重要的[135]。
6 潜在应用与展望Rieske非血红素铁环羟基化双加氧酶在多环芳烃降解中发挥着重要的作用,它是唯一能催化多环芳烃形成顺式二氢二醇的酶,与多环芳烃污染修复有着紧密关联。其一,根据RHOs的保守性,能对环境中存在的多环芳烃降解菌株进行定量及筛选,结合宏基因组学相关技术[122],能够获取到更多不可培养微生物的基因信息,不但丰富对于不可培养微生物的认识,更能够获取到新的高效降解多环芳烃的基因。其二,RHOs可以作为一种多环芳烃污染场地的指示物,针对RHOs保守区域制备相应的引物即可监测受污染场所的芳烃降解菌株的种群丰度[118]。其三,在长期污染的环境中,RHOs是可以在菌群中水平转移的,从而使得微生物能够适应这种极端恶劣环境[50]。人工模拟自然条件下的水平转移,可获得含有快速进化的高效RHOs的微生物,将其应用到污染环境中,能够更好地适应环境以降解污染物。其四,伴随着合成生物学的发展,通过分子模拟来设计和改造RHOs,结合定向突变技术[101],能够获得更多有益的RHOs的突变体,扩宽RHOs的底物催化范围,一种底物范围广、高效而且稳定的酶制剂或人工微生物对多环芳烃污染治理有着潜在的应用价值。其五,多环芳烃诸多的降解基因簇也被用于构建基因工程微生物(Genetically engineered microorganisms,GEMs),如nah-like基因簇等,GEMs可被用来清理污染场地,提高污染物的去除能力及范围[136]。目前,已经有一些RHOs或利用基因工程技术改造过的RHOs被申请专利,未来有潜力被用来处理受到有毒或致癌芳香族化合物污染的土壤、湿地和污水[137-139]。对于多环芳烃污染场地的生物修复来说,RHOs有着广阔的应用前景。
| [1] |
Jiang R, Wu X, Xiao Y, et al. Tween 20 regulate the function and structure of transmembrane proteins of Bacillus Cereus: promoting transmembrane transport of fluoranthene. J Hazard Mater, 2021, 403: 123707. DOI:10.1016/j.jhazmat.2020.123707
|
| [2] |
Patel AB, Shaikh S, Jain KR, et al. Polycyclic aromatic hydrocarbons: sources, toxicity, and remediation approaches. Front Microbiol, 2020, 11: 562813. DOI:10.3389/fmicb.2020.562813
|
| [3] |
Nzila A, Musa MM. Current status of and future perspectives in bacterial degradation of benzo[a]pyrene. Int J Environ Res Public Heal, 2020, 18(1): 262. DOI:10.3390/ijerph18010262
|
| [4] |
Menzie CA, Potocki BB, Santodonato J. Exposure to carcinogenic PAHs in the environment. Environ Sci Technol, 1992, 26(7): 1278-1284. DOI:10.1021/es00031a002
|
| [5] |
土壤环境质量建设用地土壤污染风险管控标准(试行) GB36600-2018). [2021-08-27]. http://www.mee.gov.cn/ywgz/fgbz/bz/bzwb/trhj/201807/t20180703_446027.shtml. Soil environmental quality risk control standard for soil contamination of development land (Trial) (GB36600-2018). [2021-08-27]. http://www.mee.gov.cn/ywgz/fgbz/bz/bzwb/trhj/201807/t20180703_446027.shtml (in Chinese). |
| [6] |
Abdel-Shafy HI, Mansour MSM. A review on polycyclic aromatic hydrocarbons: source, environmental impact, effect on human health and remediation. Egypt J Petroleum, 2016, 25(1): 107-123. DOI:10.1016/j.ejpe.2015.03.011
|
| [7] |
Zhang Z, Guo H, Sun J, et al. Exploration of the biotransformation processes in the biodegradation of phenanthrene by a facultative anaerobe, strain PheF2, with Fe(Ⅲ) or O2 as an electron acceptor. Sci Total Environ, 2021, 750: 142245. DOI:10.1016/j.scitotenv.2020.142245
|
| [8] |
Premnath N, Mohanrasu K, Guru Raj Rao R, et al. A crucial review on polycyclic aromatic hydrocarbons-environmental occurrence and strategies for microbial degradation. Chemosphere, 2021, 280: 130608. DOI:10.1016/j.chemosphere.2021.130608
|
| [9] |
Elufisan TO, Rodríguez-Luna IC, Oyedara OO, et al. The polycyclic aromatic hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp.. Pemsol isolated from Mexico. PeerJ, 2020, 8: e8102.
|
| [10] |
Wang P, Zhang Y, Jin J, et al. A high-efficiency phenanthrene-degrading Diaphorobacter sp. isolated from PAH-contaminated river sediment. Sci Total Environ, 2020, 746: 140455. DOI:10.1016/j.scitotenv.2020.140455
|
| [11] |
张金宝, 李凤梅, 郭书海, 等. 高分子量多环芳烃降解菌筛选及在土壤电动-生物修复中应用. 生态学杂志, 2020, 39(1): 260-269. Zhang JB, Li FM, Guo SH, et al. Isolation of high molecular weight PAHs degrading bacteria and its application in the electro-bioremediation of contaminated soil. Chin J Ecol, 2020, 39(1): 260-269 (in Chinese). |
| [12] |
何芬, 王立华, 宁大亮, 等. 嗜盐菌群对菲的降解及萘双加氧酶基因的表达规律. 中国环境科学, 2012, 32(9): 1662-1669. He F, Wang LH, Ning DL, et al. Phenanthrene biodegradation and dynamic change of expression of naphthalene dioxygenase(ndo) genes in a halophilic bacteria consortium. China Environ Sci, 2012, 32(9): 1662-1669 (in Chinese). DOI:10.3969/j.issn.1000-6923.2012.09.018 |
| [13] |
王慧, 周海燕, 黄勇, 等. 一株高环多环芳烃降解嗜盐菌Thalassospira sp.的分离及降解特性. 清华大学学报(自然科学版), 2015, 55(1): 87-92. Wang H, Zhou HY, Huang Y, et al. Isolation and degradation characteristics of a HMW-PAHs degrading halophilic strain Thalassospira sp.. J Tsinghua Univ Sci Technol, 2015, 55(1): 87-92 (in Chinese). |
| [14] |
Fu B, Xu T, Cui Z, et al. Mutation of phenylalanine-223 to leucine enhances transformation of benzo[a]pyrene by ring-hydroxylating dioxygenase of Sphingobium sp. FB3 by increasing accessibility of the catalytic site. J Agric Food Chem, 2018, 66(5): 1206-1213. DOI:10.1021/acs.jafc.7b05018
|
| [15] |
Wang WW, Li QG, Zhang LG, et al. Genetic mapping of highly versatile and solvent-tolerant Pseudomonas putida B6 -2 (ATCC BAA -2545) as a 'superstar' for mineralization of PAHs and dioxin-like compounds. Environ Microbiol, 2021, 23(8): 4309-4325. DOI:10.1111/1462-2920.15613
|
| [16] |
Liu YL, Hu HY, Zanaroli G, et al. A Pseudomonas sp. strain uniquely degrades PAHs and heterocyclic derivatives via lateral dioxygenation pathways. J Hazard Mater, 2021, 403: 123956. DOI:10.1016/j.jhazmat.2020.123956
|
| [17] |
Yu H, Li T, Liu Y, et al. Spatial distribution of polycyclic aromatic hydrocarbon contamination in urban soil of China. Chemosphere, 2019, 230: 498-509. DOI:10.1016/j.chemosphere.2019.05.006
|
| [18] |
Zhang Y, Peng C, Guo Z, et al. Polycyclic aromatic hydrocarbons in urban soils of China: distribution, influencing factors, health risk and regression prediction. Environ Pollut, 2019, 254(pt a): 112930.
|
| [19] |
Han J, Liang Y, Zhao B, et al. Polycyclic aromatic hydrocarbon (PAHs) geographical distribution in China and their source, risk assessment analysis. Environ Pollut, 2019, 251: 312-327. DOI:10.1016/j.envpol.2019.05.022
|
| [20] |
谢林培, 祝冲之, 张晓东, 等. 强化生物堆法修复多环芳烃污染土壤的初步研究. 生态与农村环境学报, 2021, 37(1): 96-102. Xie LP, Zhu CZ, Zhang XD, et al. Preliminary study on remediation of PAH-contaminated soil by enhanced biopile. J Ecol Rural Environ, 2021, 37(1): 96-102 (in Chinese). |
| [21] |
Fang T, Zhou NY. Purification and characterization of salicylate 5-hydroxylase, a three-component monooxygenase from Ralstonia sp. strain U2. Appl Microbiol Biotechnol, 2014, 98(2): 671-679. DOI:10.1007/s00253-013-4914-x
|
| [22] |
Liu TT, Xu Y, Liu H, et al. Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl Microbiol Biotechnol, 2011, 90(2): 671-678. DOI:10.1007/s00253-010-3033-1
|
| [23] |
Jin J, Yao J, Liu W, et al. Fluoranthene degradation and binding mechanism study based on the active-site structure of ring-hydroxylating dioxygenase in Microbacterium paraoxydans JPM1. Environ Sci Pollut Res Int, 2017, 24(1): 363-371. DOI:10.1007/s11356-016-7809-4
|
| [24] |
周子康, 崔洁, 许平, 等. 细菌降解低分子量多环芳烃的研究进展. 生物工程学报, 2019, 35(11): 2069-2080. Zhou ZK, Cui J, Xu P, et al. Progress in biodegradation of low molecular weight polycyclic aromatic hydrocarbons. Chin J Biotechnol, 2019, 35(11): 2069-2080 (in Chinese). |
| [25] |
吴洁婷, 许琪, 张营, 等. 微生物降解典型高分子量多环芳烃的研究进展. 环境科学研究, 2021, 34(8): 1981-1990. Wu JT, Xu Q, Zhang Y, et al. Progress in microbial degradation of typical HMW-PAHs. Research of Environmental Sciences, 2021, 34(8): 1981-1990 (in Chinese). |
| [26] |
Bugg TD, Ramaswamy S. Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations. Curr Opin Chem Biol, 2008, 12(2): 134-140. DOI:10.1016/j.cbpa.2007.12.007
|
| [27] |
Özgen FF, Schmidt S. Rieske non-heme iron dioxygenases: applications and future perspectives. Biocatalysis, 2019, 57-82.
|
| [28] |
Gally C, Nestl BM, Hauer B. Engineering rieske non-heme iron oxygenases for the asymmetric dihydroxylation of alkenes. Angew Chem Int Ed Engl, 2015, 54(44): 12952-12956. DOI:10.1002/anie.201506527
|
| [29] |
Wang CH, Lu JW, Wei HH, et al. Functional model for intradiol-cleaving catechol 1, 2-dioxygenase: synthesis, structure, spectra, and catalytic activity of iron (Ⅲ) complexes with substituted salicylaldimine ligands. Inorganica Chimica Acta, 2007, 360(9): 2944-2952. DOI:10.1016/j.ica.2007.02.034
|
| [30] |
Erwin KL, Johnson WH, Meichan AJ, et al. Preparation of dihydroxy polycyclic aromatic hydrocarbons and activities of two dioxygenases in the phenanthrene degradative pathway. Arch Biochem Biophys, 2019, 673: 108081. DOI:10.1016/j.abb.2019.108081
|
| [31] |
Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta, 2007, 1770(3): 314-329. DOI:10.1016/j.bbagen.2006.07.003
|
| [32] |
Syed K, Doddapaneni H, Subramanian V, et al. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem Biophys Res Commun, 2010, 399(4): 492-497. DOI:10.1016/j.bbrc.2010.07.094
|
| [33] |
Sutherland JB, Selby AL, Freeman JP, et al. Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl Environ Microbiol, 1991, 57(11): 3310-3316. DOI:10.1128/aem.57.11.3310-3316.1991
|
| [34] |
Chen C, Shen J, Yang L, et al. Identification of structural properties influencing the metabolism of polycyclic aromatic hydrocarbons by cytochrome P450 1A1. Sci Total Environ, 2021, 758: 143997. DOI:10.1016/j.scitotenv.2020.143997
|
| [35] |
宁大亮. 白腐真菌细胞色素P450转化难降解有机物的功能研究. 北京: 清华大学, 2009. Ning DL. Function of cytochrome P450 in degradation of refractory organic compounds by a white rot fungus. Beijing: Tsinghua University, 2009 (in Chinese). |
| [36] |
Syed K, Porollo A, Lam YW, et al. CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl Environ Microbiol, 2013, 79(8): 2692-2702. DOI:10.1128/AEM.03767-12
|
| [37] |
罗安. Rhodococcus sp. P14降解多环芳烃过程中双加氧酶Baa和细胞色素P450单加氧酶CYP108J1功能的研究. 汕头: 汕头大学, 2015. Luo A. Functional analysis of dioxygenase Baa and cytochrome P450 monoxygenase CYP108J1 involved in polycyclic aromatic hydrocarbon degradation in Rhodococcus sp. P14. Shantou: Shantou University, 2015 (in Chinese). |
| [38] |
Brezna B, Kweon O, Stingley RL, et al. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaalenii PYR-1. Appl Microbiol Biotechnol, 2006, 71(4): 522-532. DOI:10.1007/s00253-005-0190-8
|
| [39] |
Butler CS, Mason JR. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol, 1997, 38: 47-84.
|
| [40] |
Spain JC, Zylstra GJ, Blake CK, et al. Monohydroxylation of phenol and 2, 5-dichlorophenol by toluene dioxygenase in Pseudomonas putida F1. Appl Environ Microbiol, 1989, 55(10): 2648-2652. DOI:10.1128/aem.55.10.2648-2652.1989
|
| [41] |
Höring P, Rothschild-Mancinelli K, Sharma ND, et al. Oxidative biotransformations of phenol substrates catalysed by toluene dioxygenase: a molecular docking study. J Mol Catal B: Enzym, 2016, 134: 396-406. DOI:10.1016/j.molcatb.2016.10.013
|
| [42] |
Resnick S, Lee K, Gibson D. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Ind Microbiol, 1996, 17(5/6): 438-457.
|
| [43] |
Jouanneau Y, Meyer C, Jakoncic J, et al. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry, 2006, 45(40): 12380-12391. DOI:10.1021/bi0611311
|
| [44] |
Kasai Y, Shindo K, Harayama S, et al. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Appl Environ Microbiol, 2003, 69(11): 6688-6697. DOI:10.1128/AEM.69.11.6688-6697.2003
|
| [45] |
Jerina DM, Selander H, Yagi H, et al. Dihydrodiols from anthracene and phenanthrene. J Am Chem Soc, 1976, 98(19): 5988-5996. DOI:10.1021/ja00435a035
|
| [46] |
Brezna B, Khan AA, Cerniglia CE. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp.. FEMS Microbiol Lett, 2003, 223(2): 177-183. DOI:10.1016/S0378-1097(03)00328-8
|
| [47] |
Asturias JA, Diaz E, Timmis KN. The evolutionary relationship of biphenyl dioxygenase from Gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from Gram-negative bacteria. Gene, 1995, 156(1): 11-18. DOI:10.1016/0378-1119(94)00530-6
|
| [48] |
Hedlund BP, Geiselbrecht AD, Staley JT. Marinobacter strain NCE312 has a Pseudomonas-like naphthalene dioxygenase. FEMS Microbiol Lett, 2001, 201(1): 47-51. DOI:10.1111/j.1574-6968.2001.tb10731.x
|
| [49] |
Benedek T, Szentgyörgyi F, Szabó I, et al. Aerobic and oxygen-limited naphthalene-amended enrichments induced the dominance of Pseudomonas spp. from a groundwater bacterial biofilm. Appl Microbiol Biotechnol, 2020, 104(13): 6023-6043. DOI:10.1007/s00253-020-10668-y
|
| [50] |
Herrick JB, Stuart-Keil KG, Ghiorse WC, et al. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol, 1997, 63(6): 2330-2337.
|
| [51] |
Mawad AMM, Abdel-Mageed WS, Hesham AE. Quantification of naphthalene dioxygenase (NahAC) and catechol dioxygenase (C23O) catabolic genes produced by phenanthrene-degrading Pseudomonas fluorescens AH-40. Curr Genomics, 2020, 21(2): 111-118. DOI:10.2174/1389202921666200224101742
|
| [52] |
Kumari S, Mangwani N, Das S. Naphthalene catabolism by biofilm forming marine bacterium Pseudomonas aeruginosa N6P6 and the role of quorum sensing in regulation of dioxygenase gene. J Appl Microbiol, 2021, 130(4): 1217-1231. DOI:10.1111/jam.14867
|
| [53] |
Imperato V, Portillo-Estrada M, McAmmond BM, et al. Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of bóbrka (Poland). Genes, 2019, 10(6): 443. DOI:10.3390/genes10060443
|
| [54] |
Subashchandrabose SR, Venkateswarlu K, Naidu R, et al. Biodegradation of high-molecular weight PAHs by Rhodococcus wratislaviensis strain 9: overexpression of amidohydrolase induced by pyrene and BaP. Sci Total Environ, 2019, 651(pt 1): 813-821.
|
| [55] |
Baratto MC, Lipscomb DA, Larkin MJ, et al. Spectroscopic characterisation of the naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. Int J Mol Sci, 2019, 20(14): 3402. DOI:10.3390/ijms20143402
|
| [56] |
Miyazawa D, Thanh LTH, Tani A, et al. Isolation and characterization of genes responsible for naphthalene degradation from thermophilic naphthalene degrader, Geobacillus sp. JF8. Microorganisms, 2019, 8(1): E44. DOI:10.3390/microorganisms8010044
|
| [57] |
Zhao Q, Bilal M, Yue S, et al. Identification of biphenyl 2, 3-dioxygenase and its catabolic role for phenazine degradation in Sphingobium yanoikuyae B1. J Environ Manage, 2017, 204(pt 1): 494-501.
|
| [58] |
Chakraborty J, Das S. Characterization of the metabolic pathway and catabolic gene expression in biphenyl degrading marine bacterium Pseudomonas aeruginosa JP-11. Chemosphere, 2016, 144: 1706-1714. DOI:10.1016/j.chemosphere.2015.10.059
|
| [59] |
Jin JN, Yao J, Zhang QY, et al. An integrated approach of bioassay and molecular docking to study the dihydroxylation mechanism of pyrene by naphthalene dioxygenase in Rhodococcus sp. ustb-1. Chemosphere, 2015, 128: 307-313. DOI:10.1016/j.chemosphere.2015.02.012
|
| [60] |
Pham TT, Sondossi M, Sylvestre M. Metabolism of doubly para-substituted hydroxychlorobiphenyls by bacterial biphenyl dioxygenases. Appl Environ Microbiol, 2015, 81(14): 4860-4872. DOI:10.1128/AEM.00786-15
|
| [61] |
Musumeci MA, Loviso CL, Lozada M, et al. Substrate specificities of aromatic ring-hydroxylating oxygenases of an uncultured gammaproteobacterium from chronically-polluted subantarctic sediments. Int Biodeterior Biodegrad, 2019, 137: 127-136. DOI:10.1016/j.ibiod.2018.12.005
|
| [62] |
Zhang XW, Qu YY, Ma Q, et al. Cloning and expression of naphthalene dioxygenase genes from Comamonas sp. MQ for indigoids production. Process Biochem, 2013, 48(4): 581-587. DOI:10.1016/j.procbio.2013.02.008
|
| [63] |
Cui CZ, Feng TC, Yu YQ, et al. Isolation, charcaterization of an anthracene degrading bacterium Martelella sp. AD-3 and cloning of dioxygenase gene. Huan Jing Ke Xue (Chinese), 2012, 33(11): 4062-4068.
|
| [64] |
Viger JF, Mohammadi M, Barriault D, et al. Metabolism of chlorobiphenyls by a variant biphenyl dioxygenase exhibiting enhanced activity toward dibenzofuran. Biochem Biophys Res Commun, 2012, 419(2): 362-367. DOI:10.1016/j.bbrc.2012.02.029
|
| [65] |
Tittabutr P, Cho IK, Li QX. Phn and Nag-like dioxygenases metabolize polycyclic aromatic hydrocarbons in Burkholderia sp. C3. Biodegradation, 2011, 22(6): 1119-1133. DOI:10.1007/s10532-011-9468-y
|
| [66] |
Robrock KR, Mohn WW, Eltis LD, et al. Biphenyl and ethylbenzene dioxygenases of Rhodococcus jostii RHA1 transform PBDEs. Biotechnol Bioeng, 2011, 108(2): 313-321. DOI:10.1002/bit.22952
|
| [67] |
Lee TK, Lee J, Sul WJ, et al. Novel biphenyl-oxidizing bacteria and dioxygenase genes from a Korean tidal mudflat. Appl Environ Microbiol, 2011, 77(11): 3888-3891. DOI:10.1128/AEM.00023-11
|
| [68] |
Di Gennaro P, Terreni P, Masi G, et al. Identification and characterization of genes involved in naphthalene degradation in Rhodococcus opacus R7. Appl Microbiol Biotechnol, 2010, 87(1): 297-308. DOI:10.1007/s00253-010-2497-3
|
| [69] |
Fukuhara Y, Kasai D, Katayama Y, et al. Enzymatic properties of terephthalate 1, 2-dioxygenase of Comamonas sp. strain E6. Biosci Biotechnol Biochem, 2008, 72(9): 2335-2341. DOI:10.1271/bbb.80236
|
| [70] |
Patrauchan MA, Florizone C, Eapen S, et al. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Sphingobium RHA1. J Bacteriol, 2008, 190(1): 37-47. DOI:10.1128/JB.01122-07
|
| [71] |
Iwasaki T, Takeda H, Miyauchi K, et al. Characterization of two biphenyl dioxygenases for biphenyl/PCB degradation in a PCB degrader, Rhodococcus sp. strain RHA1. Biosci Biotechnol Biochem, 2007, 71(4): 993-1002. DOI:10.1271/bbb.60663
|
| [72] |
Gómez-Gil L, Kumar P, Barriault D, et al. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J Bacteriol, 2007, 189(15): 5705-5715. DOI:10.1128/JB.01476-06
|
| [73] |
Chadhain SM, Moritz EM, Kim E, et al. Identification, cloning, and characterization of a multicomponent biphenyl dioxygenase from Sphingobium yanoikuyae B1. J Ind Microbiol Biotechnol, 2007, 34(9): 605-613. DOI:10.1007/s10295-007-0235-3
|
| [74] |
Francova K, Macková M, Macek T, et al. Ability of bacterial biphenyl dioxygenases from Burkholderia sp. LB400 and Comamonas testosteroni B-356 to catalyse oxygenation of ortho-hydroxychlorobiphenyls formed from PCBs by plants. Environ Pollut, 2004, 127(1): 41-48. DOI:10.1016/S0269-7491(03)00257-4
|
| [75] |
Barriault D, Lépine F, Mohammadi M, et al. Revisiting the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase toward 2, 2'-dichlorobiphenyl and 2, 3, 2', 3'-tetrachlorobiphenyl. J Biol Chem, 2004, 279(46): 47489-47496. DOI:10.1074/jbc.M406808200
|
| [76] |
Zhou NY, Al-Dulayymi J, Baird MS, et al. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J Bacteriol, 2002, 184(6): 1547-1555. DOI:10.1128/JB.184.6.1547-1555.2002
|
| [77] |
Laurie AD, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol, 1999, 181(2): 531-540. DOI:10.1128/JB.181.2.531-540.1999
|
| [78] |
Sylvestre M, Sirois M, Hurtubise Y, et al. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene, 1996, 174(2): 195-202. DOI:10.1016/0378-1119(96)00039-X
|
| [79] |
Lau PC, Bergeron H, Labbé D, et al. Sequence and expression of the todGIH genes involved in the last three steps of toluene degradation by Pseudomonas putida F1. Gene, 1994, 146(1): 7-13. DOI:10.1016/0378-1119(94)90827-3
|
| [80] |
Tan HM, Tang HY, Joannou CL, et al. The Pseudomonas putida ML2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low in G+C content. Gene, 1993, 130(1): 33-39. DOI:10.1016/0378-1119(93)90343-2
|
| [81] |
Taira K, Hirose J, Hayashida S, et al. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem, 1992, 267(7): 4844-4853. DOI:10.1016/S0021-9258(18)42908-0
|
| [82] |
Erickson BD, Mondello FJ. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol, 1992, 174(9): 2903-2912. DOI:10.1128/jb.174.9.2903-2912.1992
|
| [83] |
Kurkela S, Lehväslaiho H, Palva ET, et al. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene, 1988, 73(2): 355-362. DOI:10.1016/0378-1119(88)90500-8
|
| [84] |
Yang JW, Cho W, Lim Y, et al. Evaluation of aromatic hydrocarbon decomposition catalyzed by the dioxygenase system and substitution of ferredoxin and ferredoxin reductase. Environ Sci Pollut Res Int, 2019, 26(33): 34047-34057. DOI:10.1007/s11356-018-3200-y
|
| [85] |
Yu CL, Liu W, Ferraro DJ, et al. Purification, characterization, and crystallization of the components of a biphenyl dioxygenase system from Sphingobium yanoikuyae B1. J Ind Microbiol Biotechnol, 2007, 34(4): 311-324. DOI:10.1007/s10295-006-0199-8
|
| [86] |
Inoue K, Usami Y, Ashikawa Y, et al. Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1, 9a-dioxygenase. Appl Environ Microbiol, 2014, 80(9): 2821-2832. DOI:10.1128/AEM.04000-13
|
| [87] |
Kounosu A, Hasegawa K, Iwasaki T, et al. Crystallization and preliminary X-ray diffraction studies of hyperthermophilic archaeal Rieske-type ferredoxin (ARF) from Sulfolobus solfataricus P1. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2010, 66(pt 7): 842-845.
|
| [88] |
Friemann R, Lee K, Brown EN, et al. Structures of the multicomponent Rieske non-heme iron toluene 2, 3-dioxygenase enzyme system. Acta Crystallogr D Biol Crystallogr, 2009, 65(pt 1): 24-33.
|
| [89] |
Miao LL, Qu J, Liu ZP. Hydroxylation at multiple positions initiated the biodegradation of indeno[1, 2, 3-cd]pyrene in Rhodococcus aetherivorans IcdP1. Front Microbiol, 2020, 11: 568381. DOI:10.3389/fmicb.2020.568381
|
| [90] |
Correll CC, Batie CJ, Ballou DP, et al. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science, 1992, 258(5088): 1604-1610. DOI:10.1126/science.1280857
|
| [91] |
Ferraro DJ, Okerlund AL, Mowers JC, et al. Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1, 2-dioxygenase. J Bacteriol, 2006, 188(19): 6986-6994. DOI:10.1128/JB.00707-06
|
| [92] |
Fabara AN, Fraaije MW. An overview of microbial indigo-forming enzymes. Appl Microbiol Biotechnol, 2020, 104(3): 925-933. DOI:10.1007/s00253-019-10292-5
|
| [93] |
Jouanneau Y, Meyer C, Duraffourg N. Dihydroxylation of four-and five-ring aromatic hydrocarbons by the naphthalene dioxygenase from Sphingomonas CHY-1. Appl Microbiol Biotechnol, 2016, 100(3): 1253-1263. DOI:10.1007/s00253-015-7050-y
|
| [94] |
Ferraro DJ, Okerlund A, Brown E, et al. One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1, 2-dioxygenase. IUCrJ, 2017, 4(Pt 5): 648-656.
|
| [95] |
Kumar P, Mohammadi M, Dhindwal S, et al. Structural insights into the metabolism of 2-chlorodibenzofuran by an evolved biphenyl dioxygenase. Biochem Biophys Res Commun, 2012, 421(4): 757-762. DOI:10.1016/j.bbrc.2012.04.078
|
| [96] |
Parales RE. The role of active-site residues in naphthalene dioxygenase. J Ind Microbiol Biotechnol, 2003, 30(5): 271-278. DOI:10.1007/s10295-003-0043-3
|
| [97] |
Kauppi B, Lee K, Carredano E, et al. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1, 2-dioxygenase. Structure, 1998, 6(5): 571-586. DOI:10.1016/S0969-2126(98)00059-8
|
| [98] |
Yu CL, Parales RE, Gibson DT. Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enantioselectivity. J Ind Microbiol Biotechnol, 2001, 27(2): 94-103. DOI:10.1038/sj.jim.7000168
|
| [99] |
Jakoncic J, Jouanneau Y, Meyer C, et al. The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. Febs J, 2007, 274(10): 2470-2481. DOI:10.1111/j.1742-4658.2007.05783.x
|
| [100] |
Ferraro DJ, Brown EN, Yu CL, et al. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2, 3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct Biol, 2007, 7: 10. DOI:10.1186/1472-6807-7-10
|
| [101] |
Li JD, Min J, Wang Y, et al. Engineering Burkholderia xenovorans LB400 BphA through site-directed mutagenesis at position 283. Appl Environ Microbiol, 2020, 86(19): e01040-20.
|
| [102] |
Barriault D, Sylvestre M. Evolution of the biphenyl dioxygenase BphA from Burkholderia xenovorans LB400 by random mutagenesis of multiple sites in region Ⅲ. J Biol Chem, 2004, 279(46): 47480-47488. DOI:10.1074/jbc.M406805200
|
| [103] |
Madej T, Lanczycki CJ, Zhang D, et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res, 2014, 42(database issue): D297-D303.
|
| [104] |
Mohammadi M, Viger JF, Kumar P, et al. Retuning Rieske-type oxygenases to expand substrate range. J Biol Chem, 2011, 286(31): 27612-27621. DOI:10.1074/jbc.M111.255174
|
| [105] |
Kumar P, Mohammadi M, Viger JF, et al. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J Mol Biol, 2011, 405(2): 531-547. DOI:10.1016/j.jmb.2010.11.009
|
| [106] |
Colbert CL, Agar NY, Kumar P, et al. Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes. PLoS ONE, 2013, 8(1): e52550. DOI:10.1371/journal.pone.0052550
|
| [107] |
Ashikawa Y, Fujimoto Z, Noguchi H, et al. Electron transfer complex formation between oxygenase and ferredoxin components in Rieske nonheme iron oxygenase system. Structure, 2006, 14(12): 1779-1789. DOI:10.1016/j.str.2006.10.004
|
| [108] |
Gakhar L, Malik ZA, Allen CC, et al. Structure and increased thermostability of Rhodococcus sp. naphthalene 1, 2-dioxygenase. J Bacteriol, 2005, 187(21): 7222-7231. DOI:10.1128/JB.187.21.7222-7231.2005
|
| [109] |
Friemann R, Ivkovic-Jensen MM, Lessner DJ, et al. Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J Mol Biol, 2005, 348(5): 1139-1151. DOI:10.1016/j.jmb.2005.03.052
|
| [110] |
Dong X, Fushinobu S, Fukuda E, et al. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J Bacteriol, 2005, 187(7): 2483-2490. DOI:10.1128/JB.187.7.2483-2490.2005
|
| [111] |
Nojiri H, Ashikawa Y, Noguchi H, et al. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1, 9a-dioxygenase. J Mol Biol, 2005, 351(2): 355-370. DOI:10.1016/j.jmb.2005.05.059
|
| [112] |
Karlsson A, Parales JV, Parales RE, et al. NO binding to naphthalene dioxygenase. JBIC J Biol Inorg Chem, 2005, 10(5): 483-489. DOI:10.1007/s00775-005-0657-1
|
| [113] |
Furusawa Y, Nagarajan V, Tanokura M, et al. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J Mol Biol, 2004, 342(3): 1041-1052. DOI:10.1016/j.jmb.2004.07.062
|
| [114] |
Karlsson A, Parales JV, Parales RE, et al. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science, 2003, 299(5609): 1039-1042. DOI:10.1126/science.1078020
|
| [115] |
Carredano E, Karlsson A, Kauppi B, et al. Substrate binding site of naphthalene 1, 2-dioxygenase: functional implications of indole binding. J Mol Biol, 2000, 296(2): 701-712. DOI:10.1006/jmbi.1999.3462
|
| [116] |
Kauppi B, Lee K, Carredano E, et al. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1, 2-dioxygenase. Structure, 1998, 6(5): 571-586. DOI:10.1016/S0969-2126(98)00059-8
|
| [117] |
Piubeli FA, dos Santos LG, Fernández EN, et al. The emergence of different functionally equivalent PAH degrading microbial communities from a single soil in liquid PAH enrichment cultures and soil microcosms receiving PAHs with and without bioaugmentation. Pol J Microbiol, 2018, 67(3): 365-375. DOI:10.21307/pjm-2018-046
|
| [118] |
Meynet P, Head IM, Werner D, et al. Re-evaluation of dioxygenase gene phylogeny for the development and validation of a quantitative assay for environmental aromatic hydrocarbon degraders. FEMS Microbiol Ecol, 2015, 91(6): fiv049.
|
| [119] |
Panicker G, Mojib N, Aislabie J, et al. Detection, expression and quantitation of the biodegradative genes in Antarctic microorganisms using PCR. Antonie Van Leeuwenhoek, 2010, 97(3): 275-287. DOI:10.1007/s10482-009-9408-6
|
| [120] |
Hedlund BP, Staley JT. Isolation and characterization of Pseudoalteromonas strains with divergent polycyclic aromatic hydrocarbon catabolic properties. Environ Microbiol, 2006, 8(1): 178-182. DOI:10.1111/j.1462-2920.2005.00871.x
|
| [121] |
Baldwin BR, Nakatsu CH, Nies L. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microbiol, 2003, 69(6): 3350-3358. DOI:10.1128/AEM.69.6.3350-3358.2003
|
| [122] |
Loviso CL, Lozada M, Guibert LM, et al. Metagenomics reveals the high polycyclic aromatic hydrocarbon-degradation potential of abundant uncultured bacteria from chronically polluted subantarctic and temperate coastal marine environments. J Appl Microbiol, 2015, 119(2): 411-424. DOI:10.1111/jam.12843
|
| [123] |
Mercadal JP, Isaac P, Siñeriz F, et al. Indigo production by Pseudomonas sp. J26, a marine naphthalene-degrading strain. J Basic Microbiol, 2010, 50(3): 290-293. DOI:10.1002/jobm.200900276
|
| [124] |
Wissner JL, Escobedo-Hinojosa W, Heinemann PM, et al. Methods for the detection and analysis of dioxygenase catalyzed dihydroxylation in mutant derived libraries. Methods Enzymol, 2020, 644: 63-93.
|
| [125] |
Ma Q, Zhang X, Qu Y. Biodegradation and biotransformation of indole: advances and perspectives. Front Microbiol, 2018, 9: 2625. DOI:10.3389/fmicb.2018.02625
|
| [126] |
Khara P, Roy M, Chakraborty J, et al. Characterization of a topologically unique oxygenase from Sphingobium sp. PNB capable of catalyzing a broad spectrum of aromatics. Enzyme Microb Technol, 2018, 111: 74-80. DOI:10.1016/j.enzmictec.2017.10.006
|
| [127] |
Gao YZ, Liu XY, Liu H, et al. A bph-like nitroarene dioxygenase catalyzes the conversion of 3-nitrotoluene to 3-methylcatechol by Rhodococcus sp. strain ZWL3NT. Appl Environ Microbiol, 2020, 86(4): e02517-19.
|
| [128] |
Preston-Herrera C, Jackson AS, Bachmann BO, et al. Development and application of a high throughput assay system for the detection of Rieske dioxygenase activity. Org Biomol Chem, 2021, 19(4): 775-784. DOI:10.1039/D0OB02412K
|
| [129] |
Fossbakk A, Haavik J. An oxygraphic method for determining kinetic properties and catalytic mechanism of aromatic amino acid hydroxylases. Anal Biochem, 2005, 343(1): 100-105. DOI:10.1016/j.ab.2005.04.043
|
| [130] |
Hurtubise Y, Barriault D, Powlowski J, et al. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J Bacteriol, 1995, 177(22): 6610-6618. DOI:10.1128/jb.177.22.6610-6618.1995
|
| [131] |
Maeda T, Takahashi Y, Suenaga H, et al. Functional analyses of Bph-Tod hybrid dioxygenase, which exhibits high degradation activity toward trichloroethylene. J Biol Chem, 2001, 276(32): 29833-29838. DOI:10.1074/jbc.M102025200
|
| [132] |
Demanèche S, Meyer C, Micoud J, et al. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl Environ Microbiol, 2004, 70(11): 6714-6725. DOI:10.1128/AEM.70.11.6714-6725.2004
|
| [133] |
Escalante DE, Aukema KG, Wackett LP, et al. Simulation of the bottleneck controlling access into a rieske active site: predicting substrates of naphthalene 1, 2-dioxygenase. J Chem Inf Model, 2017, 57(3): 550-561. DOI:10.1021/acs.jcim.6b00469
|
| [134] |
Cao YM, Xu L, Jia LY. Analysis of PCBs degradation abilities of biphenyl dioxygenase derived from Enterobacter sp. LY402 by molecular simulation. N Biotechnol, 2011, 29(1): 90-98. DOI:10.1016/j.nbt.2011.08.005
|
| [135] |
Dhindwal S, Gomez-Gil L, Neau DB, et al. Structural basis of the enhanced pollutant-degrading capabilities of an engineered biphenyl dioxygenase. J Bacteriol, 2016, 198(10): 1499-1512. DOI:10.1128/JB.00952-15
|
| [136] |
Wu C, Li F, Yi SW, et al. Genetically engineered microbial remediation of soils co-contaminated by heavy metals and polycyclic aromatic hydrocarbons: advances and ecological risk assessment. J Environ Manag, 2021, 296: 113185. DOI:10.1016/j.jenvman.2021.113185
|
| [137] |
Kasai Y, Misawa N, Shindo K. New aromatic ring dioxygenase gene cluster, useful for cleaning environment polluted with toxic or carcinogenic aromatic compounds: JPN, WO2004050875-A1. 2004-06-17.
|
| [138] |
Iida T, Kudo T. Novel dibenzofuran dioxygenase gene derived from Terrabacter genus encoding dibenzofuran dioxygenase, comprising dioxygenase alpha, beta subunit, ferredoxin and ferredoxin reductase subunit, useful in bioremediation: JPN, JP2003250569-A. 2003-09-09.
|
| [139] |
Wang Y, Ban J. Optimized dibenzofuran dioxygenase gene dfda1 for improving antifouling performance of microorganisms, used in ecological restoration of coastal wetlands and sewage treatment, has one thousand four hundred and twenty-five nucleotides: CHN, CN107384881-A. 2017-11-24.
|
 2021, Vol. 37
2021, Vol. 37




