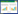中国科学院微生物研究所、中国微生物学会主办
文章信息
- 孙中贯, 刘琳, 王亚平, 王雪山, 肖冬光
- Sun Zhongguan, Liu Lin, Wang Yaping, Wang Xueshan, Xiao Dongguang
- 酿酒酵母高级醇代谢研究进展
- Higher alcohols metabolism by Saccharomyces cerevisiae: a mini review
- 生物工程学报, 2021, 37(2): 429-447
- Chinese Journal of Biotechnology, 2021, 37(2): 429-447
- 10.13345/j.cjb.200302
-
文章历史
- Received: May 28, 2020
- Accepted: August 10, 2020
- Published: September 3, 2020
2. 天津科技大学 工业发酵微生物教育部重点实验室,天津 300457;
3. 枣庄学院 食品科学与制药工程学院,山东 枣庄 277160;
4. 中国轻工业浓香型白酒固态发酵重点实验室,四川 宜宾 644000;
5. 枣庄学院 生命科学学院,山东 枣庄 277160
2. Key Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin University of Science and Technology, Tianjin 300457, China;
3. College of Food Science and Pharmaceutical Engineering, Zaozhuang University, Zaozhuang 277160, Shandong, China;
4. Key Laboratory of Wuliangye-flavor Liquor Solid-state Fermentation, China National Light Industry, Yibin 644000, Sichuan, China;
5. College of Life Science, Zaozhuang University, Zaozhuang 277160, Shandong, China
在饮料酒酿造过程中,酿酒酵母 Saccharomyces cerevisiae 除产生主要代谢产物乙醇外,还产生高级醇类、酯类、醛类、酚类、酸类、连二酮类等风味代谢产物[1]。其中,高级醇,俗称杂醇油,是具有2个以上碳链骨架的一价醇类的统称[2]。正丙醇、异丁醇、异戊醇、2-甲基-1-丁醇(活性戊醇)、2-苯乙醇(图 1) 是酿酒酵母产生的主要高级醇类物质[3];各高级醇类物质均具有自身独特的呈香呈味特征[4] (表 1),在饮料酒中高级醇类物质以不同比例存在并共同作用形成各类风味独特的饮料酒。
| Higher alcohols | Flavor characteristics |
| n-propanol | Resembles that of ethanol, but relatively dense, seems to have ether odour |
| Isobutanol | Stronger ethanol or higher alcohol odor, also have fat incense |
| Isoamyl alcohol | Typical higher alcohol aroma with an unpleasant odor |
| 2-methyl-1-butanol | Higher alcohol odor, slightly floral |
| 2-phenylalcohol | Floral odor of roses |
适量的高级醇能够赋予饮料酒丰满的口感,柔和协调的酒体;但其含量过高时,极易给饮料酒带来异杂味,饮用后易产生口渴、头痛等症状,对人体产生明显的副作用,不利于饮用者的身体健康[2, 10-11]。饮料酒中正丙醇含量过高,易产生似醚臭、有苦味;丁醇含量过高,易产生杂醇油臭味[12];戊醇超标则有腐败味和汗臭味[13];2-苯乙醇接近阈值时,有酯类的酸味[14]。有研究报道,高级醇能够抑制人体的神经中枢,对交感神经和视觉神经等产生损伤。相同浓度下,高级醇的麻醉作用要强于乙醇;若乙醇的麻醉作用为1,则丙醇为8.5,异丁醇为8,异戊醇为19;此外,人体分解高级醇的速度也较慢,导致体内高级醇的代谢停留时间较长[15]。这些因素也是导致饮用高级醇含量过高的饮料酒时,醉酒较慢且醉酒后较难醒酒的主要原因[11]。目前对于饮料酒中高级醇类物质的含量并没有形成统一的标准,大量的调查研究表明,下面发酵啤酒中高级醇适宜含量应小于100 mg/L,优质啤酒低于50 mg/L;葡萄酒中高级醇最适含量不宜超过300 mg/L。黄酒中为80–540 mg/L,小曲液态发酵白酒中为600–2 500 mg/L,固态发酵白酒中为500–1 800 mg/L,白兰地中为1 000–2 000 mg/L,朗姆酒中为650–2 000 mg/L,威士忌中为500–1 500 mg/L;优质饮料酒中一般高级醇含量较低,而劣质饮料酒中普遍存在高级醇含量过高的问题。
高级醇属于初级代谢产物,是在酵母菌的生长繁殖过程中形成的;因而其主要形成时期为饮料酒主发酵期,一般在3–7 d结束。在酒精发酵后期,发酵前期生成的醇类物质和酸类物质在酶的催化作用下生成酯类物质。在多数情况下,后发酵期持续时间越长,酯类物质的含量越高,成品酒的品质也就越好。这正是发酵周期较短的中低档饮料酒醇高酯低的主要原因之一,也是名优酒都采用较长发酵周期最主要的原因。然而,发酵周期的延长将导致酒损大、效率低、成本高,特别是高档白酒原料出酒率仅为理论出酒率的30%–50%。由此可见,探明酿酒酵母高级醇类物质的代谢机理,完善高级醇类物质代谢网络,实现高级醇类物质代谢途径的精细化调控,对降低酿酒工业粮耗、缩短发酵周期、改善饮料酒品质具有重要意义。
本文系统总结了酿酒酵母合成高级醇类物质的代谢途径以及诱变育种与代谢工程技术在高级醇代谢调控中的应用,重点介绍了代谢工程技术改造高级醇类物质代谢途径的研究思路。最后,本文展望了实现酿酒酵母高级醇类物质代谢途径精细化调控的未来发展趋势。
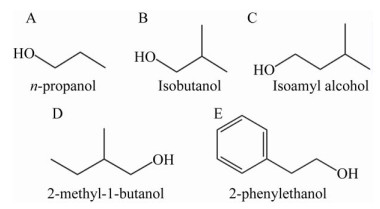
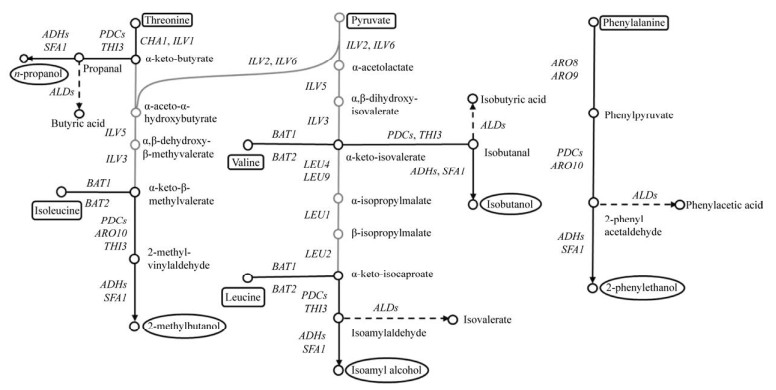
1 酿酒酵母合成高级醇的代谢途径及主要影响因素酿酒酵母利用发酵原料中游离的氨基氮合成自身生长繁殖所需的蛋白质,当氨基酸中的氨基被利用后,残余的α-酮酸经脱羧和加氢还原可生成相应的高级醇[16],即氨基酸分解代谢途径;这一代谢途径最初是由德国化学家Felix Ehrlich提出的,也被称为Ehrlich途径[2-3] (图 2)。此后,Harris研究糖代谢时发现,丙酮酸经支链氨基酸合成代谢途径可生成α-酮酸,进而生成高级醇,即丙酮酸合成代谢途径[2-3] (图 2)。
|
| 图 2 酿酒酵母高级醇合成代谢网络图[17] Fig. 2 Biosynthetic pathways for higher alcohols formation in S. cerevisiae[17]. Black solid lines represent that the higher alcohols are derived from the Ehrlich pathway. Gray solid lines represent the synthesis of corresponding amino acid. Black dotted lines represent the synthesis of corresponding carboxylic acids. |
| |
1904年,德国化学家Ehrlich在研究氨基酸和高级醇的化学结构式时,发现亮氨酸与异戊醇(3-甲基-1-丁醇)、异亮氨酸与2-甲基-1-丁醇的化学结构存在相似性,Ehrlich由此猜测氨基酸通过分解代谢形成了高级醇,同时生成一分子的氨基并释放一分子的CO2。为证明这一猜想,Ehrlich在酿酒酵母发酵培养基中添加氨基酸,实验结果显示高级醇的生成量显著增加[1, 4]。1911年,Neubauer和Fromherz提出了α-酮酸为氨基酸分解代谢的第一个中间产物,α-酮酸经脱羧反应生成醛,醛加氢还原生成高级醇,形成了沿用至今的Ehrlich途径。随后,科学家们通过研究证实在酿酒酵母代谢过程中,所有的高级醇均可由相应的氨基酸分解产生[2]。经过一个多世纪的研究,Ehrlich途径不断被完善,现在可以总结为:氨基酸在氨基转移酶的催化作用下生成α-酮酸,同时将氨基转移给α-酮戊二酸生成谷氨酸,α-酮酸在脱羧酶的作用下生成相应的醛和CO2;醛经加氢还原生成相应的醇。
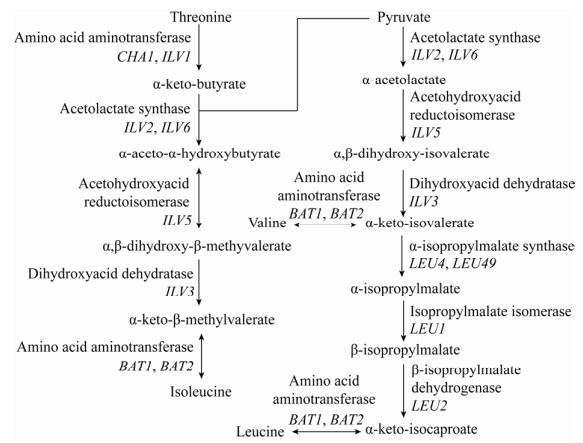
1.2 丙酮酸合成代谢途径酿酒酵母利用丙酮酸生成的α-酮酸,可经脱羧和还原反应生成高级醇,也可与氨基发生合成反应生成相应的氨基酸。酵母菌在利用丙酮酸合成氨基酸的过程中,若氨基供应不足,将导致过量的α-酮酸生成相应的高级醇。现已研究证实,丙酮酸可通过支链氨基酸合成代谢途径(Isoleucine-leucine- valine biosynthesis pathway,ILV pathway) 实现异亮氨酸、亮氨酸、缬氨酸的生物合成(图 3)。
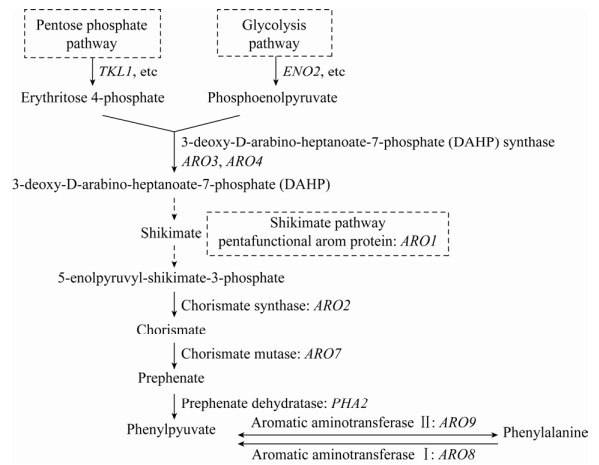
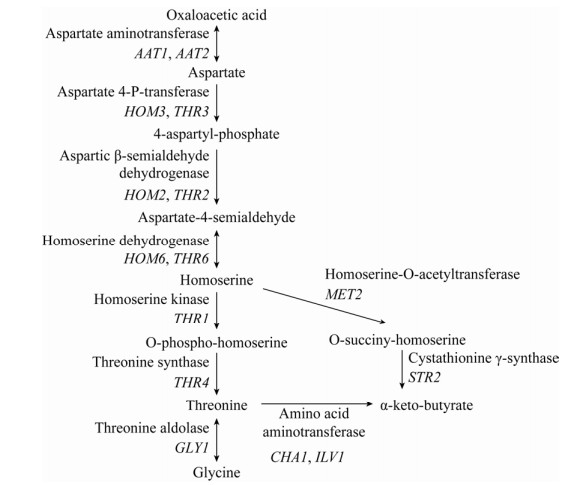
酿酒酵母也可利用莽草酸途径(Shikimate pathway) 实现苯丙酮酸的从头合成,来自糖酵解途径的磷酸烯醇式丙酮酸与来自磷酸戊糖途径的4-磷酸赤藓糖经多步酶促反应后可生成莽草酸,莽草酸再经多步酶促反应即可生成苯丙酮酸(图 4)。三羧酸循环中的草酰乙酸与氨基发生加成反应生成天冬氨酸,天冬氨酸经过5步酶促反应即可生成苏氨酸,此外,苏氨酸和甘氨酸也可在苏氨酸醛缩酶的催化下实现相互转化(图 5);这两条代谢途径是合成α-酮丁酸的主要来源。
1.3 两条代谢途径对高级醇合成的贡献目前,在酿酒酵母细胞中合成代谢途径和分解代谢途径对高级醇合成的贡献尚不明朗。有研究表明,饮料酒中的异丁醇、异戊醇和2-甲基-1-丁醇总量的75%来自合成代谢途径,25%来自分解代谢途径[20];但这一比例与发酵液中可游离氨基氮含量有密切关系。通常情况下分解代谢途径在发酵早期进行,此时发酵液中游离氨基氮含量较高;而在发酵后期,若游离氨基氮被酵母彻底利用,合成代谢途径将被激活[21]。
当发酵液中游离氨基氮过量,尤其为氨基酸种类及浓度充足时,酵母细胞不需要自身合成氨基酸,同时氨基酸对合成代谢途径具有反馈抑制作用,酵母细胞则主要通过分解代谢途径合成高级醇。当可利用发酵液中游离氨基氮匮乏时(如高辅料比麦芽汁),由于氨基的供应量不足导致酵母代谢产生的α-酮酸无法生成相应的氨基酸,此时α-酮酸则主要生成高级醇[21]。Jiang等利用Box-Behnken设计试验并结合响应面分析发现,碳氮比对白酒酿造过程中高级醇的生成具有显著的影响[22]。由此可见,发酵原料中碳氮源的含量与种类以及碳氮源的比例等,是影响两条高级醇合成代谢途径活跃程度的关键因素。
2 酿酒酵母菌株改造策略通过发酵原料和发酵工艺的优化调控酿酒酵母的生长代谢活动,从而间接控制高级醇的生成量是一条简捷方便的途径。但由于饮料酒发酵原料和发酵工艺的独特性和复杂性,通过优化发酵原料和发酵工艺的方式调控酿酒酵母高级醇的生成量存在较大困难。因而,关注酿酒酵母菌种、选育出具有适宜高级醇生成量的优良菌株是一条最为有效的途径。目前,研究人员主要采用诱变育种和代谢工程技术调控酿酒酵母高级醇代谢活动,通过选育氨基酸营养缺陷型菌株或改变与高级醇代谢途径相关的某些基因表达量的方式,达到有效控制高级醇生成量的目的。
2.1 诱变育种对亲本菌株进行诱变处理和筛选是改良菌株性状的有效手段之一,其中目的菌株的高通量筛选是诱变育种的关键环节。诱变育种技术在酿酒酵母高级醇代谢调控中的应用已有较多的报道(表 2)。秦伟帅利用甲基磺酸乙酯对二倍体葡萄酒酵母菌株S. cerevisiae EC1118进行诱变处理并结合有性重组技术,在含氯乙酸异戊酯的合成基础(Synthetic dropout,SD) 培养基上筛选获得一株高级醇生成量较出发菌株降低了14.1%的突变菌株[20]。王国正等利用常温常压等离子体诱变和筛选得到一株高级醇生成量比亲本菌株S. cerevisiae CF4降低了20.0%的突变株[23]。本课题组是较早利用人工诱变育种技术选育低产高级醇酿酒酵母菌株的课题组之一。韩涛对啤酒酵母S. cerevisiae SC-2进行紫外线诱变处理,以亮氨酸营养缺陷型为筛选标记,通过制霉菌素法淘汰野生型菌株,最终获得一株高级醇生成量降低25.5%的亮氨酸营养缺陷型菌株[24]。王鹏银等通过低能氮离子注入诱变和制霉菌素法淘汰野生型,获得一株具有亮氨酸营养缺陷特性的突变株。与亲本菌株S. cerevisiaeAY-15相比,该突变株的高级醇生成总量降低33.6%,异戊醇生成量降低39.9%[25]。郑玲艳等以清酒酵母S. cerevisiae JS-10为出发菌株,利用紫外线诱变技术结合重氮染色平板筛选,选育出一株高级醇生成量下降32.7%的酵母菌株[26]。在饮料酒酿造过程中,由于生产原料营养丰富,普遍存在高级醇生成量过高的现象,但也存在因酵母菌种、发酵条件等原因造成高级醇生成量不足的问题。为改善单一酵母菌种导致黄酒风味物质含量不足的问题,Yang等利用乙醇定向驯化、紫外线诱变处理筛选克霉唑抗性突变株以及原生质体融合等方法,获得一株乙醇耐受性显著增强同时高级醇合成水平提高13.4%–24.91%的黄酒酵母S. cerevisiae F23[27]。Tian等为提高高酸性条件下青梅果酒酵母S. cerevisiae ET008的高级醇合成水平,利用常温常压等离子体对出发菌株进行诱变,以生长速度为筛选标记,96孔板培养法进行初筛;对初筛获得的120株突变株,以风味物质含量为筛选标记,6孔板培养法进行复筛;对5株风味物质生成量最高的突变株进行实验室适应性驯化,驯化70代后,最终获得一株耐酸能力强、高级醇合成量提高38.2%的突变株[28]。
| Strains | Alcoholic beverages | Mutation breeding | Change in total higher alcohol | References |
| EC1118 | Wine | Ethyl methanesulfonate and protoplast fusion | 237.4 mg/L, decreased by 14.1% | [21] |
| CF4 | Chinese Baijiu | Atmospheric and room temperature plasma (ARTP) | 66.4 mg/L, decreased by 20.0% | [23] |
| SC-2 | Lager beer | UV | 94.5 mg/L, decreased by 25.5% | [24] |
| AY-15 | Chinese Baijiu | Ion implantation | 482.6 mg/L, decreased by 33.6% | [25] |
| JS-10 | Sake | UV | 90.7 mg/L, decreased by 32.7% | [26] |
| BR20 and BR30 | Chinese Huangjiu | Ethanol domestication, ultraviolet radiation (UV) and protoplast fusion | 51.85 mg/L, increased by 13.4%–24.9% | [27] |
| ET008 | Greengage fruit wine | ARTP, high-throughput screening (HTS), and adaptive laboratory evolution (ALE) | 3.1 mg/L, increased by 38.2% | [28] |
| YDZ | Lager beer | UV | 67.9 mg/L, decreased by 28.7% | [29] |
| HF2.3 | Lager beer | UV and diethyl sulfate | 70.7 mg/L, decreased by 24.3% | [30] |
酿酒酵母能够利用乳酸脱氢酶催化α-酮酸生成乳酸。乳酸脱氢酶活性的提高,可实现α-酮酸的竞争性利用,从而达到降低高级醇生成量的目的。汪志君等利用紫外线对啤酒酵母S. cerevisiae YZD进行诱变,通过2, 3, 5-三苯基氯化四氮唑上层平板、乳酸等筛选条件,得到一株高级醇合成水平下降28.7%的优质酵母菌株[29]。赵辉等利用紫外线和硫酸二乙酯联合诱变的方式对酿酒酵母S. carlsbergensis HF2.3进行处理,以生长速度为筛选标记,依次利用乳酸培养基、麦芽汁碳酸钙培养基、2, 3, 5-氯化三苯基四氮唑培养基进行筛选,选育出一株高乳酸脱氢酶活性的酵母菌株,其高级醇生成量较出发菌株降低24.3%[30]。
综上所述,利用诱变处理的方式能够有效降低酿酒酵母菌株的高级醇合成能力。人工诱变育种技术为饮料酒生产提供了大量具有应用潜力的优质菌株,也为利用全基因组学、转录组学、蛋白质组学等组学技术研究酿酒酵母高级醇代谢调控网络提供了理想的实验菌株。
2.2 代谢工程育种随着酿酒酵母S. cerevisiae S288c全基因组序列的公开以及高级醇代谢途径研究的不断深入,运用代谢工程技术定向改造高级醇代谢途径成为了研究的热点。目前的研究报道主要集中在氨基转移酶编码基因、α-酮酸合成代谢基因、α-酮酸分解代谢基因、乙酸酯代谢基因、碳氮代谢基因等高级醇代谢相关基因的遗传改造方面。
2.2.1 氨基转移酶编码基因的定向改造氨基转移酶能够催化氨基酸脱去氨基生成α-酮酸,是Ehrlich途径中的第一步反应。依据所催化氨基酸种类的不同,可将氨基转移酶分为两类,一类是由BAT1和BAT2基因编码的支链氨基酸氨基转移酶,另一类是由ARO8和ARO9基因编码的芳香族氨基酸氨基转移酶。此外,由CHA1和ILV1基因编码的苏氨酸氨基裂解酶具有催化苏氨酸脱去氨基的功能。
酿酒酵母产生的高级醇中主体成分为由支链氨基酸转化而成的异丁醇、异戊醇和2-甲基-1-丁醇,因而目前的研究多集中于BAT基因(表 3)。由表 3可知,敲除BAT1和BAT2基因均能够有效降低酿酒酵母高级醇的合成水平,单独过量表达BAT2基因能够有效提高酿酒酵母高级醇合成水平。Eden等以酵母粉-蛋白胨-葡萄糖(Yeast extract peptone dextrose,YPD) 培养基为培养基质,研究BAT1和BAT2基因的敲除对酿酒酵母S. cerevisiae MD101高级醇代谢的影响时发现,单独敲除BAT1和BAT2基因均能够有效降低高级醇的生成量[31]。本课题组研究证实单独敲除白酒酵母S. cerevisiae AY15和工业黄酒酵母S. cerevisiae RY1的BAT2基因以及组合敲除啤酒酵母S. cerevisiae S6和工业黄酒酵母S. cerevisiae RY1两种单倍体的BAT1和BAT2基因均能显著降低亲本菌株的高级醇合成能力[17, 36-37];然而,Eden等发现同时敲除酵母S. cerevisiae MD101的BAT1和BAT2基因,高级醇的生成量却显著提升[31]。Styger等研究发现过量表达酵母菌株S. cerevisiae BY4741的BAT1基因导致亲本菌株的高级醇合成水平显著降低,当培养基中添加过量支链氨基酸时重组菌株的高级醇合成水平与亲本菌株无显著差异[32]。Lilly等以葡萄酒酵母工业菌株S. cerevisiae VIN13为亲本菌株,发现分别过量表达BAT1和BAT2基因均能有效提高亲本菌株高级醇的生成能力,但过量表达BAT2基因的提高幅度较大[33]。Colón等已通过研究证实BAT2基因编码的酿酒酵母细胞质中的支链氨基酸氨基转移酶,负责催化支链氨基酸生成相应的α-酮酸;而BAT1基因编码的酿酒酵母线粒体中的支链氨基酸氨基转移酶,则负责催化α-酮酸生成相应的氨基酸[40]。本课题组研究发现敲除BAT2基因同时过量表达BAT1基因,能够显著降低酵母菌株的高级醇合成能力[38-39]。BAT1基因的遗传改造所引起的不同酵母菌株间的差异化表现,可能与培养基质以及菌株遗传背景有较大关系。Hammer等研究发现敲除BAT1基因能够提高酿酒酵母S. cerevisiae CEN. PK2-1C和S. cerevisiae BY4741的异丁醇合成能力;在此基础上减少缬氨酸的供给,能够进一步显著促进丙酮酸向α-乙酰乳酸的转化,进而通过支链氨基酸合成代谢途径促进异丁醇的生成[41],这项研究表明BAT1基因的敲除导致酿酒酵母缬氨酸合成能力下降,同时激活了支链氨基酸合成代谢途径;研究结果为揭示BAT基因在酿酒酵母高级醇代谢调控中的作用提供了新的研究思路。
| Strategy | Strains | Medium | Change in total higher alcohol | References |
| Deletion of BAT1 | S6 | Wort (18 °P) | 166.0 mg/L, decreased by 5.5% | [17] |
| MD101 | YPD (4% Glu) | Decreased by 20.0%–30.0% | [31] | |
| BY4742 | SCD5 (5% Glu) | 77.2 mg/L, decreased by 62.1% | [32] | |
| BY4742 | SCD5+++ (5% Glu, 150 mg/L each BCAAs) | 185.3 mg/L, decreased by 24.3% | [32] | |
| Overexpression of BAT1 | BY4742 | SCD5 (5% Glu) | 140.2 mg/L, decreased by 31.1% | [32] |
| BY4742 | SCD5+++ (5% Glu, 150 mg/L each BCAAs) | 248.7 mg/L, hardly improved | [32] | |
| VIN13 | Colombard grape juice | 223.1 mg/L, increased by 20.1% | [33] | |
| VIN13 | Synthetic must MS300 | Increased by 50.0%–60.0% | [34] | |
| Deletion of BAT2 | S6 | Wort (18 °P) | 159.5 mg/L, decreased by 9.2% | [17] |
| MD101 | YPD (4% Glu) | Decreased by 60.0%–70.0% | [31] | |
| BY4742 | SCD5 (5% Glu) | 99.1 mg/L, decreased by 51.3% | [32] | |
| BY4742 | SCD5+++ (5% Glu, 150 mg/L each BCAAs) | 145.9 mg/L, decreased by 40.4% | [32] | |
| TD4 | YPD (10% Glu) | 154.3 mg/L, decreased by 43.8% | [35] | |
| AY15 | Corn hydrolysate | 235.3 mg/L, decreased by 30.5% | [36] | |
| RY1 | Rice mash | Decreased by 10.0%–20.0% | [37] | |
| Overexpressionof BAT2 | BY4742 | SCD5 (5% Glu) | 254.2 mg/L, increased by 51.3% | [32] |
| BY4742 | SCD5+++ (5% Glu, 150 mg/L each BCAAs) | 474.3 mg/L, increased by 93.8% | [32] | |
| VIN13 | Colombard grape juice | 254.8 mg/L, increased by 37.2% | [33] | |
| TD4 | YPD (10% Glu) | 410.5 mg/L, increased by 49.6% | [35] | |
| Deletion of BAT1 and BAT2 | S6 | Wort (18 °P) | 142.1 mg/L, decreased by 19.1% | [17] |
| MD101 | PD (4% Glu) | Increased by 30.0%–40.0% | [31] | |
| RY1-a1 | Rice mash | Decreased by 31.2% | [37] | |
| RY1-α3 | Rice mash | Decreased by 28.3% | [37] | |
| Deletion of BAT2 and overexpression of BAT1 | AY15-α5 | Corn mash | Decreased by 20.0%–30.0% | [38] |
| YZ22 | Cabernet Sauvignon grape juice | 292.8 mg/L, decreased by 36.9% | [39] |
具有玫瑰香气的2-苯乙醇,目前已被广泛应用于食品、化妆品和药品等领域;酿酒酵母作为生产2-苯乙醇的优势菌株,成为了研究的热点(表 4)。Dickinson等利用13C同位素标记与核磁共振检测技术首次揭示了酿酒酵母利用苯丙氨酸合成2-苯乙醇的代谢途径[46]。苯丙氨酸能够在ARO8基因编码的组成型芳香族氨基酸氨基转移酶Ⅰ或ARO9基因编码的诱导型芳香族氨基酸氨基转移酶Ⅱ的催化作用下,发生转氨基反应生成苯丙酮酸[47]。当培养基中含有谷氨酰胺、天冬酰胺和无机氮源等偏好性氮源时,ARO9基因的表达受到抑制;当ARO8基因无法表达或培养基中仅存在非偏好性氮源时,苯丙氨酸、色氨酸、甲硫氨酸等芳香族氨基酸将诱导ARO9基因的表达[48-49],此外,研究证实敲除酿酒酵母单倍体菌株的ARO8基因,能够显著提高酵母菌株的2-苯乙醇合成水平[42-43];当同时完全敲除ARO8和ARO9基因后,酵母菌株无法完成苯丙氨酸的转氨基反应[50]。然而,有学者研究发现过量表达ARO8或ARO9基因,也能够提高2-苯乙醇的产量[44-45]。这可能是因为ARO8基因的缺失诱导了ARO9基因的表达[48-49],从而提高了酿酒酵母的2-苯乙醇合成水平;而ARO8基因主要参与苯丙氨酸的脱氨基反应[47],因而该基因的过量表达同样能够提高2-苯乙醇的合成水平。这些研究成果为揭示ARO8和ARO9基因在酿酒酵母高级醇代谢中的功能奠定了一定的研究基础,提供了可借鉴的研究思路,然而ARO8和ARO9基因参与2-苯乙醇代谢的调控机制仍需要大量的研究加以论证。
| Strategy | Strains | Medium | Change in 2-phenylethanol | References |
| Deletion of ARO8 | CEN.PK113-7D | Glu SM (2.0% Glu, ammonium as sole nitrogen source) | Detectable, about 60.0–70.0 mg/L | [42] |
| BY4741 | SD minimal medium (2.0% Glu) | Increased by 1.2–1.3 fold | [43] | |
| Overexpression of ARO8 | S288c | Fermentation media I (8.0% Glu, 0.7% of L-phenylalanine as the sole nitrogen source) | Increased by 9.3% | [44] |
| Overexpression of ARO9 | W303-1B | SC (2.0% Glu, without leucine and uracil) | Increased by 160.0%–170.0% | [45] |
正丙醇在饮料酒中的含量一般为5–25 mg/L,且风味特征不突出,因而有关ILV1和CHA1基因的研究也相对较少。本课题组Li等通过实验证实完全敲除白酒酵母S. cerevisiae AY15的ILV1基因,其正丙醇的合成水平下降30.3%[51]。
2.2.2 α-酮酸合成代谢基因的定向改造α-酮酸是酿酒酵母合成高级醇的关键中间代谢产物,其含量对高级醇的合成起到非常重要的作用。通过对丙酮酸合成代谢途径关键基因以及调控因子的表达进行调控,调节酵母细胞α-酮酸的合成水平,达到定向调控高级醇生成量的目的(表 5)。
| Strains | Medium | Strain breeding | Change in total higher alcohol | References |
| JAy1 | Minimal medium (10.0% Glu, 1×YNB) | Overexpression of ILV2, ILV3 and ILV5 | 183.0 mg/L, increased by 2.3 fold, isobutanol, 136.0 mg/L, increased by 3.9 fold | [52] |
| CEN.PK 2-1C | Minimal medium (4.0% glucose and 0.01% uracil) | Overexpression of ILV2, ILV3 and ILV5 | Isobutanol, increased by 5.0 fold | [53] |
| CEN.PK 2-1C | Minimal medium (4.0% glucose and 0.01% uracil) | Overexpression of ILV2, ILV3, ILV5 and ILV6 | Isobutanol, increased by 2.0 fold | [53] |
| D452-2 with kivd over-expressed | YPD medium (4.0% Glu) | Overexpression of ILV2, ILV3 and ILV5 | Isobutanol, 151.0 mg/L, increased by 62.4% | [54] |
| CEN.PK 2-1C with BAT1 and ALD6 deleted | SC-His-Trp-Uramedium (2% Glu) | Overexpression of ILV2, ILV3 and ILV5, deletion of BAT1 and ALD6 | Isobutanol, 72.1 mg/L, increased by 17.8% | [55] |
| AY15 | YEPD (16.0% Glu) | Deletion of LEU1 | Isoamyl alcohol, decreased by 33.7%, isobutanol, increased by 41.7% | [51] |
| AY15-a8 | High-concentration corn mash | Deletion of LEU1 | n-propanol, 61.8 mg/L, increased by 47.0%, isobutanol, 206.6 mg/L, increased by 158.0%, isoamyl alcohol, 97.8 mg/L, decreased by 51.0% | [56] |
| AY15 | YEPD (16.0% Glu) | Deletion of LEU2 | Isoamyl alcohol, decreased by 28.7%, isobutanol, increased by 52.2% | [51] |
| S-6 | Wort (10°P) | Deletion of LEU2 | 71.9 mg/L, decreased by 10.0%, isoamyl alcohol, 58.3 mg/L, decreased by 11.8% | [57] |
| N85-Na | Chinese Huangjiu | Deletion of LEU2 | Isoamyl alcohol, decreased by 16.2%, isobutanol, almost invariable | [58] |
| BY4741 with ARO10 and ADH7 over-expressed | SC (3.6% Gal, 0.4% Glu, additional valine, leucine, isoleucine, and uracil dropped out) | Overexpression of ILV2, ILV5, ILV3, LEU9, LEU1 and LEU2 | 1088.1 mg/L, increased by 500.0 fold | [59] |
| BY4741 with ARO10, ADH7, ILV2, ILV5, ILV3, LEU9, LEU1 and LEU2 over-expressed | SC (3.6% Gal, 0.4% Glu, additional histidine, valine, leucine, isoleucine, and uracil dropped out) | Expression of the LEU9c-ILV3c fusion protein | Isoamyl alcohol, 522.8 mg/L, increased by 89.5%, isobutanol, 540.3 mg/L, decreased by 27.3% | [60] |
| CEN.PK113-7D | Glu SM (2.0% Glu, ammonium as sole nitrogen source) | Deletion of ARO3, ARO8 and TYR1, ARO4 replaced by ARO4K229L, ARO7 replaced by ARO7G141S | 2-phenylethanol, detectable | [47] |
| AY15-α5 | Corn mash | Deletion of THR4 | n-propanol, increased by 2.6 fold | [61] |
参与丙酮酸合成代谢途径的基因主要为ILVs和LEUs基因;ILVs基因主要参与缬氨酸与异亮氨酸的合成,LEUs基因主要参与亮氨酸的合成。目前的研究多集中在通过过量表达ILVs基因提高高级醇合成水平以及通过敲除LEUs基因降低高级醇的合成水平。Avalos等研究发现组合过量表达ILV2、ILV5、ILV3基因的酿酒酵母S. cerevisiae JAy1的高级醇合成水平提高了约2.3倍,其中异丁醇的生成量提升幅度最大,提高了3.8倍[51]。过量表达ILVs基因能够显著提高酿酒酵母的高级醇合成能力,尤其是异丁醇的合成能力[52-55],而与异丁醇代谢途径相同的2-甲基-1-丁醇的生成量并没有发生显著变化[52];这种现象可能是由于培养基中苏氨酸含量相对匮乏导致α-酮丁酸供应不足所造成的,也可能与酶对底物的选择性催化作用有关。
本课题组李维等研究报道白酒酵母菌株S. cerevisiae AY15的LEU1基因缺失后,异丁醇的生成量提高了41.7%,异戊醇的生成量降低了33.7%;LEU2基因缺失后,异丁醇的生成量提高了52.2%,异戊醇的生成量降低了28.7%[51]。佐一含等单敲除工业啤酒酵母S. cerevisiae S-6的LEU2基因后,异戊醇生成量降低了11.8%[57]。LEU1与LEU2基因的敲除能够提高异丁醇的生成量,同时降低异戊醇的生成量[51, 56-58];但变化幅度因菌种特性和发酵工艺而异。
过量表达ILVs和LEUs基因,同样能够提高酿酒酵母的高级醇合成能力;此外,通过调控α-酮酸在线粒体和细胞质中的含量,可实现各类高级醇合成量的差异化调控,对改善饮料酒的风味具有积极的指导作用。Yuan等以过量表达ARO10与ADH7基因的酿酒酵母S. cerevisiae BY4741为出发菌株,过量表达ILV2、ILV5、ILV3、LEU9、LEU1以及LEU2基因,重组菌株的高级醇合成水平较出发菌株提高了约500倍;进一步过量表达线粒体2-异丙基苹果酸(2-isopropylmalate,α-IPM) 转运蛋白编码基因OAC1以提高细胞质中α-IPM的含量,异戊醇的生成量提高了20.0%,异丁醇的生成量降低了26.6%,高级醇生成总量没有发生显著变化[59]。Yuan等以酿酒酵母S. cerevisiae BY4741为出发菌株,在细胞质中重构2-异丙基苹果酸合成途径同时过量表达ARO10与ADH7基因的基础上,在细胞质中利用人工蛋白支架构建ILV3基因编码的二羟基酸脱水酶和LEU9基因编码的2-异丙基苹果酸合成酶的融合蛋白;与未构建融合蛋白的重组菌株相比,异戊醇的生成量增加了89.5%,异丁醇的生成量降低了27.3%,而高级醇生成总量没有发生显著变化[60]。这项研究结果表明,通过调控高级醇代谢途径中关键酶在酿酒酵母亚细胞中的定位也可调节各类高级醇的合成比例,为饮料酒中高级醇含量和配比的精细化调控提供了新的理论依据。
目前,有关莽草酸途径和草酰乙酸-天冬氨酸- 苏氨酸代谢途径的研究还鲜有报道。Romagnoli等研究发现组合敲除S. cerevisiae CEN. PK113-7D的ARO8、TYR1及ARO3基因同时分别利用基因ARO4K229L和ARO7G141S替代ARO4与ARO7基因以解除底物反馈抑制现象,能够大幅度提高2-苯乙醇的得率[42]。本课题组石钰等研究发现苏氨酸合酶编码基因THR4的敲除导致酵母菌株S. cerevisiae AY15单倍体α5的正丙醇生成量提高了2.6倍,并未达到降低正丙醇生成量的目的[61]。这些研究为阐明正丙醇和2-苯乙醇的调控机制提供了一定的理论依据,为全面揭示正丙醇和2-苯乙醇的代谢调控体系指明了方向。
2.2.3 α-酮酸分解代谢基因的定向改造PDC1、PDC5、PDC6、ARO10、THI3等基因编码的脱羧酶催化α-酮酸脱羧形成醛类物质,同时产生二氧化碳;醛类物质在ADH1、ADH2、ADH3、ADH4、ADH5、ADH6、ADH7、SFA1等基因编码的醇脱氢酶的催化作用下加氢还原生成相应的醇。醛类物质也可以在ALD2、ALD3、ALD4、ALD5、ALD6等基因编码的醛脱氢酶的催化下脱氢氧化生成相应的酸。然而,高级醇的代谢途径并不完全相同,2-甲基-1-丁醇的前体物质2-甲基-乙烯醛无法经过脱氢反应生成相应的酸;此外,脱羧酶编码基因和醇脱氢酶编码基因在各高级醇代谢途径中的作用也存在着差异[1]。由代谢调控理论可知,降低脱羧酶和醇脱氢酶的活性同时提高醛脱氢酶的活性,可以达到降低高级醇生成量的目的;反之亦然(表 6)。
| Strains | Medium | Strain breeding | Change in total higher alcohol | References |
| BY4742 | SCD5 (5.0% Glu) | Deletion of HOM2 | Decreased by 62.5% | [32] |
| BY4742 | SCD5+++ (5.0% Glu, 150 mg/L each BCAAs) | Deletion of HOM2 | Decreased by 49.7% | [32] |
| BY4742 | SCD5 (5.0% Glu) | Deletion of PAD1 | Decreased by 57.3% | [32] |
| BY4742 | SCD5+++ (5.0% Glu, 150 mg/L each BCAAs) | Deletion of PAD1 | Decreased by 26.3% | [32] |
| BY4742 | SCD5 (5.0% Glu) | Deletion of QCR2 | Decreased by 52.9% | [32] |
| BY4742 | SCD5+++ (5.0% Glu, 150 mg/L each BCAAs) | Deletion of QCR2 | Decreased by 32.4% | [32] |
| BY4742 | SCD5 (5.0% Glu) | Deletion of SPE1 | Decreased by 52.9% | [32] |
| BY4742 | SCD5+++ (5.0% Glu, 150 mg/L each BCAAs) | Deletion of SPE1 | Decreased by 26.0% | [32] |
| VIN13 | Synthetic must MS300 | Overexpression of AAD10 | Increased by 85.0%–95.0% | [34] |
| VIN13 | Synthetic must MS300 | Overexpression of AAD14 | Increased by 40.0%–50.0% | [34] |
| BY4741 | SD selective medium (2.0% Glu) | Overexpression of ARO10 | 2-phenylethanol, increased by 6.0 fold | [48] |
| W303-1B with ARO9, ARO10 and ARO80 over-expressed | SC (2.0% Glu, without leucine, tryptophan and uracil) | Deletion of ALD3 | 2-phenylethanol, increased by 40.0% | [50] |
| JAy1 with ILV2, ILV3 and ILV5 over-expressed | Minimal medium (10.0% Glu, 1×YNB) | Overexpression of ARO10 and adhARE1 (Lactococcus lactis) | Increased by 5.0 fold | [52] |
| D452-2 | YPD medium (4.0% Glu) | Overexpression of kivd (Lactococcus lactis) | Isobutanol, 93.0 mg/L, increased by 3.2 fold | [54] |
| N85-Na | Rice mash | Deletion of THI3 | Almost invariable | [58] |
| BY4742 | SCD5 (5.0% Glu) | Deletion of HOM2, PRO2 and AAD6 | Isoamyl alcohol, decreased by 65.0% | [62] |
| BY4742 | SCD5 (5.0% Glu) | Deletion of BAT2, THI3 and AAD6 | Isobutanol, decreased by 75.0% | [62] |
| AY-15(a-8 and α-22) | Corn hydrolysate | Deletion of THI3 | Almost invariable | [63] |
由表 6可知,通过过量表达或敲除α-酮酸分解代谢途径中的基因能够有效调控酿酒酵母高级醇的合成水平[34-35, 53-54],特别是异丁醇、异戊醇和2-苯乙醇的合成水平[48, 50, 62];此外,酶及其催化底物在细胞中所处位置的差异也会对高级醇的合成水平产生影响。Avalos等以酿酒酵母S. cerevisiae JAy1为原始菌株,在组合过量表达ILV2、ILV5、ILV3基因的基础上,过量表达ARO10和ADH7基因,重组菌株的异丁醇生成量没有显著的变化;而将构建的过表达体系转移到线粒体中以解除酶与底物的位置差异,异丁醇的生成量较出发菌株提高了5.0倍[52]。
值得关注的是,α-酮酸分解代谢途径中的基因在调控高级醇代谢时也表现出了丰富的多样性[35]和不确定性;李童等在黄酒酵母S. cerevisiae N85尿嘧啶缺陷型单倍体菌株Na中敲除THI3基因后,发现菌株的发酵性能及高级醇合成水平均未发生明显的变化[58];本课题组郝欣等研究THI3基因的缺失对白酒酵母S. cerevisiae AY-15的a型和α型单倍体菌株发酵性能及高级醇合成水平的影响时,也得到了同样的结果[63]。
2.2.4 乙酸酯代谢基因的定向改造在饮料酒酿造过程中,高级醇类物质形成以后,绝大部分以高级醇的形式存在于饮料酒中;少部分可以和乙酰辅酶A反应生成相应的乙酸酯类化合物,ATF1、ATF2、Lg-ATF1基因编码的醇乙酰基转移酶能够催化反应的进行。同时,乙酸酯类化合物也可在IAH1基因编码的水解酶催化作用下进行水解反应生成相应的高级醇。因此,过量表达醇乙酰基转移酶编码基因或敲除IAH1基因能够达到降低酿酒酵母高级醇合成水平的目的[32, 64-69] (表 7)。
| Strains | Medium | Strain breeding | Change in higher alcohols and acetates | References |
| TD4 with BAT2 deleted | YPD (10.0% Glu) | Overexpression of ATF1 | Isobutanol, 17.7 mg/L, decreased by 74.5%, isoamyl alcohol, 82.2 mg/L, decreased by 52.5%, isoamyl acetate, 20.1 mg/L, increased by 3.7 fold, ethyl acetate, 112.4 mg/L, increased by 3.4 fold | [32] |
| VIN13 | Colombard grape juice | Overexpression of ATF1 | 121.9 mg/L, decreased by 34.4%, ethyl acetate, 533.0 mg/L, increased by 4.6 fold, isoamyl acetate, 44.6 mg/L, increased by 3.5 fold | [64] |
| VIN13 | Colombard grape juice | Overexpression of ATF2 | 170.2 mg/L, decreased by 8.4%, ethyl acetate, 92.5 mg/L, almost invariable, isoamyl acetate, 12.7 mg/L, increased by 27.0% | [64] |
| VIN13 | Colombard grape juice | Overexpression of IAH1 | 184.5 mg/L, almost invariable, ethyl acetate, 52.8 mg/L, decreased by 44.7%, isoamyl acetate, 0.64 mg/L, decreased by 93.6% | [64] |
| RY1 | Rice mash | Overexpression of ATF1, deletion of IAH1 | Isoamyl alcohol, 156.4 mg/L, decreased by 49.0%, ethyl acetate, 468.9 mg/L, increased by 20.9 fold, isoamyl acetate, 99.9 mg/L, detectable | [65] |
| RY1 | Rice mash | Overexpression of ATF2, deletion of IAH1 | Isoamyl alcohol, 257.4 mg/L, decreased by 15.6%, ethyl acetate, 137.8 mg/L, increased by 3.9 fold, isoamyl acetate, 26.7 mg/L, detectable | [66] |
| RY1 | Rice mash | Overexpression of Lg-ATF1, deletion of IAH1 | Isoamyl alcohol, 281.5 mg/L, decreased by 7.7%, ethyl acetate, 70.9 mg/L, increased by 1.5 fold, isoamyl acetate, 8.7 mg/L, detectable | [67] |
| CLX14 | Corn hydrolysate | Overexpression of ATF1 | Ethyl acetate, 78.8 mg/L, increased by 2.1 fold | [68] |
| S5 | Wort (10 °P) | Overexpression of ATF1, deletion of BAT2 | Isoamyl alcohol, 41.3 mg/L, decreased by 49.0%, ethyl acetate, 117.4 mg/L, increased by 10.5 fold, isoamyl acetate, 9.3 mg/L, detectable | [69] |
| BY4742 | YPGluc (8.0% Glu) | Deletion of ATF1 | 162.0 mg/L, almost invariable, isoamyl acetate, 0.22 mg/L, decreased by 83.9%, ethyl acetate, 13.0 mg/L, decreased by 37.5% | [70] |
| BY4742 | YPGluc (8.0% Glu) | Deletion of ATF2 | 154.8 mg/L, almost invariable, isoamyl acetate, 1.1 mg/L, decreased by 17.5%, ethyl acetate, 18.2 mg/L, decreased by 12.5% | [70] |
| AY15-α5 | Corn mash | Deletion of IAH1 | Almost invariable | [71] |
Lilly等发现在葡萄酒酵母S. cerevisiae VIN13中过量表达ATF1基因能够显著提高乙酸乙酯、乙酸异戊酯的生成量,同时高级醇的生成量明显下降;而过量表达ATF2基因对乙酸乙酯、乙酸异戊酯以及高级醇的含量影响较小[64]。Zhang等在乙酸异戊酯生成量未达到检出水平的黄酒酵母S. cerevisiae RY1中过量表达ATF1基因同时敲除IAH1基因,异戊醇的生成量降低了49.0%,乙酸乙酯的生成量提高了20.9倍;乙酸异戊酯的生成量达到99.9 mg/L[65];过量表达ATF2基因同时敲除IAH1基因,异戊醇的生成量降低15.6%,乙酸乙酯生成量提高3.9倍,乙酸异戊酯生成量达到26.7 mg/L[66];过量表达Lg-ATF1基因同时敲除IAH1基因,乙酸乙酯合成能力提高了1.5倍,乙酸异戊酯的生成量达到8.7 mg/L,异戊醇的生成量降低了7.7%[67]。Verstrepen等以酿酒酵母S. cerevisiae BY4742为亲本菌株,发现敲除ATF1基因后亲本菌株乙酸乙酯生成量下降37.5%,乙酸异戊酯生成量下降83.9%,高级醇的生成量没有发生显著变化;敲除ATF2基因后亲本菌株乙酸乙酯生成量下降12.5%,乙酸异戊酯生成量下降17.5%,高级醇的生成量没有发生显著变化[70]。由以上研究结果可知,ATF1基因的表达水平对乙酸酯的生成量影响最为显著,其次为ATF2基因,Lg-ATF1基因的影响程度最小。
通过过量表达醇乙酰基转移酶编码基因同时敲除IAH1基因能够有效提升酿酒酵母合成乙酸酯的能力,同时减弱高级醇的合成能力,这对改善饮料酒中高级醇与酯类物质的比例具有积极的意义;然而单独对IAH1基因进行遗传改造时,往往无法得到同样的效果。Lilly等在葡萄酒酵母S. cerevisiae VIN13中过量表达IAH1基因,重组菌株乙酸乙酯、乙酸异戊酯的生成量明显下降,然而高级醇的含量基本保持不变[64]。本课题组Li等构建的IAH1基因缺失重组菌株α5-IAH1的高级醇和乙酸酯生成能力与亲本菌株S. cerevisiae AY15单倍体α5相比无显著差异[71]。
2.2.5 碳氮代谢基因的定向改造酿酒酵母合成高级醇的氨基酸分解代谢途径属于氮代谢途径的一部分,丙酮酸合成代谢途径属于碳代谢的一部分,因而酿酒酵母对碳氮源的摄取能力以及代谢能力必将会对高级醇的合成水平产生影响(表 8)。研究表明,利用代谢工程改造酿酒酵母糖酵解途径、丙酮酸代谢途径、三羧酸循环、氨基酸摄取能力等碳、氮代谢活动能够有效调控高级醇合成水平[34, 72-77]。Rossouw等通过实验证明过量表达葡萄酒酵母S. cerevisiae VIN13的乙酰辅酶A合成酶亚型编码基因ACS1能够小幅提升亲本菌株的高级醇生成量[34]。本课题组孙中贯等研究发现敲除上面发酵啤酒酵母S. cerevisiae S17的非特异性氨基酸转运蛋白编码基因GAP1,能够显著降低出发菌株的高级醇合成水平[77]。由此可见,研究碳氮源代谢对高级醇合成水平的影响,探明高级醇代谢在碳氮源代谢中的作用和地位,对阐明酿酒酵母高级醇代谢调控体系具有重要意义。
| Strains | Medium | Strain breeding | Change in total higher alcohol | References |
| VIN13 | Synthetic must MS300 | Overexpression of ACS1 | Increased by 10.0%–20.0% | [34] |
| CEN.PK2-1C with the resultant n-butanol pathway | SCD medium | Deletion of GPD1, GPD2, ADH1, ADH4 and MLS1 | n-butanol, increased by more than 3.0 fold | [72] |
| CEN.PK2-1C | SC-His medium (0.083 g/L His) | Deletion of BAT1, ALD6 and LPD1 | Isobutanol, 112.6 mg/L, increased by 6.1 fold | [73] |
| CEN.PK2-1C | SC-His medium (0.083 g/L His) | Deletion of BAT1, ALD6 and LPD1, overexpression of LEU3, ILV2, ILV5, ILV3, ARO10, ADH2, MPC1 and MPC3 | Isobutanol, 330.9 mg/L, increased by 21.0 fold | [73] |
| W303-1A with BAT2, ILV2 and ILV3 over- expressed, PDC6 deleted | SC medium | Overexpression of ZWF1 | Isobutanol, 283.0 mg/L, increased by 82.6% | [74] |
| YS58 | Phe medium (3.0% Glu, 0.55% g of L-Phe) | Overexpression of CAT8, deletion of MIG1 | 2-phenylethanol, 3.6 g/L, increased by 55.0% | [75] |
| YPH499 | YPD (0.2% Leu) | Site-directed mutation of Rsp5 ubiquitin ligase | Isoamyl alcohol, increased by 2.0 to 3.0 fold | [76] |
| S17 | Wheat wort (12 °P) | Deletion of GAP1 | 210.6 mg/L, decreased by 22.0% | [77] |
自1904年德国化学家Felix Ehrlich提出酿酒酵母的高级醇合成途径以来,关于酿酒酵母高级醇代谢的研究已经历一个多世纪之久。目前酿酒酵母的高级醇代谢途径已梳理清楚,代谢途径中的酶系及其编码基因已基本明确,但对酿酒酵母菌株的改造仍存在基因功能不明确、调控效果不理想、需要引入外源基因等诸多问题。
为实现酿酒酵母工业菌株高级醇代谢的精细化调控,从宏观层面分析,应做到以下几点:第一,研究要立足于实际生产过程。由于酿酒酵母高级醇代谢调控体系表现出较强的菌种和培养条件特异性对饮料酒生产条件下工业酵母菌株各高级醇的合成规律加以研究,可更加准确地调控高级醇的代谢活动,同时为生产活动提供理论指导。第二,培养条件的研究要系统全面。发酵原料、温度、溶解氧、菌种用量等是影响高级醇代谢的重要因素,此外发酵液酸度、渗透压、氧化还原电势以及菌种互作等也是需要考虑的因素。第三,要关注酵母菌代谢活动的整体性。酿酒酵母的高级醇代谢涉及氨基酸代谢和丙酮酸代谢,而这些代谢活动都与碳氮代谢密切相关。因此,研究酿酒酵母的高级醇代谢调控体系关键在于理清饮料酒酿造条件下的碳氮代谢调控机制。从微观层面分析,针对高级醇代谢的遗传改造要注意以下几个方面。第一,采用转录组、代谢组等组学技术对调控高级醇代谢的关键基因进行深度挖掘,可实现高级醇代谢的简洁高效调控。第二,关键基因调控机制的研究应尽量全面细致。单个基因调控机制的研究,应同时构建基因敲除和过量表达重组菌株并结合重组菌株在不同营养条件下代谢流的变化,研究该基因在高级醇代谢途径中的功能。第三,饮料酒中高级醇含量的调控,要兼顾饮料酒的风味特征,不应单纯增加或降低高级醇的含量。第四,可利用基因无痕敲除技术、梯度启动子无缝插入技术等基因编辑技术,在不引入存在安全隐患的外源基因的基础上实现高级醇代谢的精细化调控,构建可用于工业生产的工业酵母优良菌株。第五,酿酒酵母作为生产高级醇等生物燃料的优势菌株,除关注高级醇的代谢调控机制外,还应加强对高级醇外排机制和分离纯化技术体系的研究。
综上所述,实现酿酒酵母高级醇代谢的精细化调控是一项复杂的系统工程,仅研究高级醇的代谢途径很难完成。研究酿酒酵母所处的外部环境条件和与之所对应的内部生理生化特征,在此基础上建立高级醇代谢调控系统和定向育种技术体系,最终达到酿酒酵母风味物质比例协调的目标。
| [1] |
Hazelwood LA, Daran JM, van Maris AJA, et al. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol, 2008, 74(8): 2259-2266. DOI:10.1128/AEM.02625-07
|
| [2] |
Pires EJ, Teixeira JA, Brányik T, et al. Yeast: the soul of beer's aroma—a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl Microbiol Biotechnol, 2014, 98(5): 1937-1949. DOI:10.1007/s00253-013-5470-0
|
| [3] |
Choi YJ, Lee J, Jang YS, et al. Metabolic engineering of microorganisms for the production of higher alcohols. mBio, 2014, 5(5): e01524-14.
|
| [4] |
李维青. 闲话醇香. 酿酒科技, 2008(10): 121-125. Li WQ. Discussion on alcohols' aroma. Liquor- making Sci Technol, 2008(10): 121-125 (in Chinese). |
| [5] |
National Center for Biotechnology Information. PubChem Substance Database[EB/OL]. [2020-05-20]. https://pubchem.ncbi.nlm.nih.gov/substance/377643250.
|
| [6] |
National Center for Biotechnology Information. Pub Chem Substance Database[EB/OL]. [2020-05-20]. https://pubchem.ncbi.nlm.nih.gov/substance/319064603.
|
| [7] |
National Center for Biotechnology Information. Pub Chem Substance Database[EB/OL]. [2020-05-20]. https://pubchem.ncbi.nlm.nih.gov/substance/319084034.
|
| [8] |
National Center for Biotechnology Information. Pub Chem Substance Database[EB/OL]. [2020-05-20]. https://pubchem.ncbi.nlm.nih.gov/substance/363906754.
|
| [9] |
National Center for Biotechnology Information. Pub Chem Substance Database[EB/OL]. [2020-05-20]. https://pubchem.ncbi.nlm.nih.gov/substance/319064063.
|
| [10] |
Procopio S, Qian F, Becker T. Function and regulation of yeast genes involved in higher alcohol and ester metabolism during beverage fermentation. Eur Food Res Technol, 2011, 233(5): 721-729. DOI:10.1007/s00217-011-1567-9
|
| [11] |
肖冬光, 陈叶福, 张翠英. 啤酒生产中高级醇的形成与低产高级醇酵母菌种的选育. 山东轻工业学院学报(自然科学版), 2013, 27(3): 15-19. Xiao DG, Chen YF, Zhang CY. Synthesis of higher alcohols during beer fermentation and brewing yeast improving for low higher alcohols producing. J Shandong Polytechn Univ (Nat Sci Ed), 2013, 27(3): 15-19 (in Chinese). |
| [12] |
Kuroda K, Ueda M. Cellular and molecular engineering of yeast Saccharomyces cerevisiae for advanced biobutanol production. FEMS Microbiol Lett, 2016, 363(3): fnv247. DOI:10.1093/femsle/fnv247
|
| [13] |
崔云前, 叶国超, 魏丽培, 等. 降低上面发酵小麦啤酒头痛感的酿造条件优化. 中国酿造, 2016, 35(5): 134-138. Cui YQ, Ye GC, Wei LP, et al. Optimization of top-fermented wheat beer brewing technology to reduce headache. China Brew, 2016, 35(5): 134-138 (in Chinese). |
| [14] |
于爱红, 耿靖玮, 弭孝涛, 等. 控制上面发酵小麦啤酒中高级醇含量的研究进展. 中国酿造, 2011(1): 21-25. Yu AH, Geng JW, Mi XT, et al. Research progress on higher alcohols control in top-fermentation wheat beer production. China Brew, 2011(1): 21-25 (in Chinese). DOI:10.3969/j.issn.0254-5071.2011.01.007 |
| [15] |
杨小兰, 罗正明, 胡仕屏, 等. 降低高浓啤酒发酵中高级醇含量的研究. 食品科学, 2011, 32(9): 188-192. Yang XL, Luo ZM, Hu SP, et al. Reducing total higher alcohol content in high gravity fermentation of beer. Food Sci, 2011, 32(9): 188-192 (in Chinese). |
| [16] |
El-Dalatony MM, Saha S, Govindwar SP, et al. Biological conversion of amino acids to higher alcohols. Trends Biotechnol, 2019, 37(8): 855-869. DOI:10.1016/j.tibtech.2019.01.011
|
| [17] |
孙中贯, 周波, 王孟祺, 等. 高温高浓发酵工业啤酒酵母菌种的构建. 生物工程学报, 2019, 35(3): 522-534. Sun ZG, Zhou B, Wang MQ, et al. Construction of industrial brewing yeast for fermentation under high temperature and high gravity condition. Chin J Biotech, 2019, 35(3): 522-534 (in Chinese). |
| [18] |
陈先锐, 王肇悦, 何秀萍. 酵母菌合成2-苯乙醇的研究进展. 生物工程学报, 2016, 32(9): 1151-1163. Chen XR, Wang ZY, He XP. Advances in biosynthesis of 2-phenylethanol by yeasts. Chin J Biotech, 2016, 32(9): 1151-1163 (in Chinese). |
| [19] |
Kyoto Encyclopedia of Genes and Genomes. KEGG PATHWAY Database[EB/OL]. [2020-05-20]. https://www.kegg.jp/kegg-bin/highlight_pathway?scale=1.0&map=sce00260.
|
| [20] |
秦伟帅. 葡萄酒酵母遗传操作构建高级醇低产菌株的研究[D]. 泰安: 山东农业大学, 2013. Qin WS. Construction of wine yeast strain with low-yield higher alcohols by genetic manipulation[D]. Tai'an: Shandong Agricultural University, 2013 (in Chinese). |
| [21] |
雷宏杰. 高浓麦汁氮源组成对酵母氨基酸同化及发酵调控影响的研究[D]. 广州: 华南理工大学, 2014. Lei HJ. Study of the effects of nitrogen composition in high gravity wort on the assimilation of amino acids by lager yeast and fermentation control[D]. Guangzhou: South China University of Technology, 2014 (in Chinese). |
| [22] |
Jiang J, Liu YC, Li HH, et al. Modeling and regulation of higher alcohol production through the combined effects of the C/N ratio and microbial interaction. J Agric Food Chem, 2019, 67(38): 10694-10701. DOI:10.1021/acs.jafc.9b04545
|
| [23] |
王国正, 吴群, 徐岩. 低产高级醇酿酒酵母突变菌株的差异蛋白组分析及高级醇合成相关蛋白的差异表达. 微生物学通报, 2015, 42(12): 2407-2416. Wang GZ, Wu Q, Xu Y. Comparative intracellular proteomics analysis of a low higher alcohols producing Saccharomyces cerevisiae mutant and different expression of higher alcohols synthesis related proteins. Microbiol China, 2015, 42(12): 2407-2416 (in Chinese). |
| [24] |
韩涛. 啤酒酿造中高温发酵低产杂醇油酵母菌种的选育[D]. 天津: 天津科技大学, 2004. Han T. Breeding of beer yeast with low yield of fusel alcohols at high temperature[D]. Tianjin: Tianjin University of Science and Technology, 2004 (in Chinese). |
| [25] |
王鹏银, 郝欣, 郭学武, 等. 离子注入诱变选育低产高级醇酿酒酵母菌株. 酿酒科技, 2008(2): 17-21, 26. Wang PY, Hao X, Guo XW, et al. Screening of saccharomyces cerevisiae strains with low yield of higher alcohols by ion implantation. Liquor-Making Sci Technol, 2008(2): 17-21, 26 (in Chinese). |
| [26] |
郑玲艳, 杨建刚, 郭学武, 等. 低产异戊醇清酒酵母菌株的选育. 酿酒科技, 2008(10): 17-19. Zheng LY, Yang JG, Guo XW, et al. Breeding selection of sake yeast strains with low-yield of isoamyl alcohol. Liquor-Making Sci Technol, 2008(10): 17-19 (in Chinese). |
| [27] |
Yang YJ, Xia YJ, Lin XN, et al. Improvement of flavor profiles in Chinese rice wine by creating fermenting yeast with superior ethanol tolerance and fermentation activity. Food Res Int, 2018, 108: 83-92. DOI:10.1016/j.foodres.2018.03.036
|
| [28] |
Tian TT, Wu DH, Ng CT, et al. A multiple-step strategy for screening Saccharomyces cerevisiae strains with improved acid tolerance and aroma profiles. Appl Microbiol Biotechnol, 2020, 104(7): 3097-3107. DOI:10.1007/s00253-020-10451-z
|
| [29] |
汪志君, 高庆, 方维明, 等. 紫外诱变筛选低高级醇和双乙酰含量的啤酒酵母. 中国酿造, 2005, 24(1): 13-17. Wang ZJ, Gao Q, Fang WM, et al. Screening of low higher-alcohols and diacetyl producing Saccharomyces cerevisiae swain with ultraviolet mutagenesis. China Brew, 2005, 24(1): 13-17 (in Chinese). DOI:10.3969/j.issn.0254-5071.2005.01.004 |
| [30] |
赵辉, 蔺善喜, 王葳, 等. 低高级醇啤酒酵母的选育及中试发酵. 食品工业科技, 2011, 32(10): 242-244, 367. Zhao H, Lin SX, Wang W, et al. Screening of Saccharomgces carlsbergensis with lower higher alcohols and pilot fermentation. Sci Technol Food Ind, 2011, 32(10): 242-244, 367 (in Chinese). |
| [31] |
Eden A, Van Nedervelde L, Drukker M, et al. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol, 2001, 55(3): 296-300. DOI:10.1007/s002530000506
|
| [32] |
Styger G, Jacobson D, Bauer FF. Identifying genes that impact on aroma profiles produced by Saccharomyces cerevisiae and the production of higher alcohols. Appl Microbiol Biotechnol, 2011, 91(3): 713-730. DOI:10.1007/s00253-011-3237-z
|
| [33] |
Lilly M, Bauer FF, Styger G, et al. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res, 2006, 6(5): 726-743. DOI:10.1111/j.1567-1364.2006.00057.x
|
| [34] |
Rossouw D, Naes T, Bauer FF. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics, 2008, 9: 530. DOI:10.1186/1471-2164-9-530
|
| [35] |
Yoshimoto H, Fukushige T, Yonezawa T, et al. Genetic and physiological analysis of branched-chain alcohols and isoamyl acetate production in Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 2002, 59(4/5): 501-508.
|
| [36] |
张艳英, 肖冬光, 张翠英, 等. BAT2缺失酿酒酵母基因工程安全菌株的构建及其杂交育种. 食品与发酵工业, 2012, 38(1): 36-40. Zhang YY, Xiao DG, Zhang CY, et al. Construction and crossing trial of BAT2 deletion mutants lacking antibiotic resistant gene in Saccharomyces cerevisiae. Food Ferment Ind, 2012, 38(1): 36-40 (in Chinese). |
| [37] |
Zhang CY, Qi YN, Ma HX, et al. Decreased production of higher alcohols by Saccharomyces cerevisiae for Chinese rice wine fermentation by deletion of bat aminotransferases. J Ind Microbiol Biotechnol, 2015, 42(4): 617-625. DOI:10.1007/s10295-015-1583-z
|
| [38] |
刘芳志, 张翠英, 李维, 等. BAT 基因改造对酿酒酵母高级醇生成量的影响. 现代食品科技, 2016, 32(6): 142-147. Liu FZ, Zhang CY, Li W, et al. Effects of BAT genetic modification on the yield of higher alcohols from Saccharomyces cerevisiae. Mod Food Sci Technol, 2016, 32(6): 142-147 (in Chinese). |
| [39] |
Ma LJ, Huang SY, Du LP, et al. Reduced production of higher alcohols by saccharomyces cerevisiae in red wine fermentation by simultaneously overexpressing BAT1 and Deleting BAT2. J Agric Food Chem, 2017, 65(32): 6936-6942. DOI:10.1021/acs.jafc.7b01974
|
| [40] |
Colón M, Hernández F, López K, et al. Saccharomyces cerevisiae Bat1 and Bat2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme. PLoS ONE, 2011, 6(1): e16099. DOI:10.1371/journal.pone.0016099
|
| [41] |
Hammer SK, Avalos JL. Uncovering the role of branched-chain amino acid transaminases in Saccharomyces cerevisiae isobutanol biosynthesis. Metab Eng, 2017, 44: 302-312. DOI:10.1016/j.ymben.2017.10.001
|
| [42] |
Romagnoli G, Knijnenburg TA, Liti G, et al. Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast, 2015, 32(1): 29-45.
|
| [43] |
Shen L, Nishimura Y, Matsuda F, et al. Overexpressing enzymes of the Ehrlich pathway and deleting genes of the competing pathway in Saccharomyces cerevisiae for increasing 2-phenylethanol production from glucose. J Biosci Bioeng, 2016, 122(1): 34-39. DOI:10.1016/j.jbiosc.2015.12.022
|
| [44] |
Yin S, Zhou H, Xiao X, et al. Improving 2-phenylethanol production via Ehrlich pathway using genetic engineered Saccharomyces cerevisiae Strains. Curr Microbiol, 2015, 70(5): 762-767. DOI:10.1007/s00284-015-0785-y
|
| [45] |
Kim B, Cho BR, Hahn JS. Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol Bioeng, 2014, 111(1): 115-124. DOI:10.1002/bit.24993
|
| [46] |
Dickinson JR, Salgado LEJ, Hewlins MJE. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem, 2003, 278(10): 8028-8034. DOI:10.1074/jbc.M211914200
|
| [47] |
Iraqui I, Vissers S, Cartiaux M, et al. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferasesⅠandⅡreveals a new aminotransferase subfamily. Mol Gen Genet, 1998, 257(2): 238-248. DOI:10.1007/s004380050644
|
| [48] |
Urrestarazu A, Vissers S, Iraqui I, et al. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet, 1998, 257(2): 230-237. DOI:10.1007/s004380050643
|
| [49] |
Boer VM, Tai SL, Vuralhan Z, et al. Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose- limited chemostat cultures. FEMS Yeast Res, 2007, 7(4): 604-620. DOI:10.1111/j.1567-1364.2007.00220.x
|
| [50] |
Pirkov I, Norbeck J, Gustafsson L, et al. A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. FEBS J, 2008, 275(16): 4111-4120. DOI:10.1111/j.1742-4658.2008.06552.x
|
| [51] |
Li W, Chen SJ, Wang JH, et al. Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation. Appl Microbiol Biotechnol, 2018, 102(4): 1783-1795. DOI:10.1007/s00253-017-8715-5
|
| [52] |
Avalos JL, Fink GR, Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol, 2013, 31(4): 335-341. DOI:10.1038/nbt.2509
|
| [53] |
Chen X, Nielsen KF, Borodina I, et al. Increased isobutanol production in Saccharomyces Cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels, 2011, 4: 21. DOI:10.1186/1754-6834-4-21
|
| [54] |
Lee WH, Seo SO, Bae YH, et al. Isobutanol production in engineered Saccharomyces cerevisiae by overexpression of 2-ketoisovalerate decarboxylase and valine biosynthetic enzymes. Bioprocess Biosyst Eng, 2012, 35(9): 1467-1475. DOI:10.1007/s00449-012-0736-y
|
| [55] |
Park SH, Kim SJ, Hahn JS. Metabolic engineering of Saccharomyces cerevisiae for the production of isobutanol and 3-methyl-1-butanol. Appl Microbiol Biotechnol, 2014, 98(21): 9139-9147. DOI:10.1007/s00253-014-6081-0
|
| [56] |
石钰, 陈叶福, 肖冬光. LEU1基因缺失对酿酒酵母高级醇生成量的影响. 酿酒科技, 2015(2): 12-16. Shi Y, Chen YF, Xiao DG. Effects of LEU1 gene deletion on higher alcohols yield of S. cerevisiae. Liquor-Making Sci Technol, 2015(2): 12-16 (in Chinese). |
| [57] |
佐一含, 朱旭东, 陈叶福, 等. LEU2基因敲除对工业啤酒酵母高级醇生成量的影响. 中国酿造, 2011(3): 27-30. Zuo YH, Zhu XD, Chen YF, et al. Effect of LEU2 gene knockout on higher alcohols production in industrial Saccharomyces cerevisiae. China Brew, 2011(3): 27-30 (in Chinese). DOI:10.3969/j.issn.0254-5071.2011.03.008 |
| [58] |
李童, 孙军勇, 吴殿辉, 等. YDL080C 和 LEU2 基因敲除对工业黄酒酵母异戊醇生成量的影响. 食品工业科技, 2015, 36(15): 189-193. Li T, Sun JY, Wu DH, et al. Effect of YDL080C and LEU2 gene knockout on isoamyl alcohol production in industrial yellow rice wine yeast. Sci Technol Food Ind, 2015, 36(15): 189-193 (in Chinese). |
| [59] |
Yuan JF, Mishra P, Ching CB. Engineering the leucine biosynthetic pathway for isoamyl alcohol overproduction in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol, 2017, 44(1): 107-117. DOI:10.1007/s10295-016-1855-2
|
| [60] |
Yuan JF, Chen X, Mishra P, et al. Metabolically engineered Saccharomyces cerevisiae for enhanced isoamyl alcohol production. Appl Microbiol Biotechnol, 2017, 101(1): 465-474. DOI:10.1007/s00253-016-7970-1
|
| [61] |
石钰, 陈叶福, 郭学武, 等. 酿酒酵母苏氨酸合成酶缺失对高级醇生成量的影响. 酿酒科技, 2014(7): 26-30. Shi Y, Chen YF, Guo XW, et al. Effects of THR4 gene deletion in S. cerevisiae on higher alcohols yield. Liquor-Making Sci Technol, 2014(7): 26-30 (in Chinese). |
| [62] |
Styger G, Jacobson D, Prior BA, et al. Genetic analysis of the metabolic pathways responsible for aroma metabolite production by Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 2013, 97(10): 4429-4442. DOI:10.1007/s00253-012-4522-1
|
| [63] |
郝欣, 肖冬光, 张翠英. 酿酒酵母类丙酮酸脱羧酶基因缺失对高级醇生成量的影响. 微生物学报, 2010, 50(8): 1030-1035. Hao X, Xiao DG, Zhang CY. Effect of YDL080C gene deletion on higher alcohols production in Saccharomyces cerevisiae haploids. Acta Microbiol Sin, 2010, 50(8): 1030-1035 (in Chinese). |
| [64] |
Lilly M, Bauer FF, Lambrechts MG, et al. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast, 2006, 23(9): 641-659. DOI:10.1002/yea.1382
|
| [65] |
Zhang JW, Zhang CY, Dai LH, et al. Effects of overexpression of the alcohol acetyltransferase-encoding gene ATF1 and disruption of the esterase-encoding gene IAH 1 on the flavour profiles of Chinese yellow rice wine. Int J Food Sci Technol, 2012, 47(12): 2590-2596. DOI:10.1111/j.1365-2621.2012.03140.x
|
| [66] |
Zhang JW, Zhang CY, Qi YN, et al. Acetate ester production by Chinese yellow rice wine yeast overexpressing the alcohol acetyltransferase-encoding gene ATF2. Genet Mol Res, 2014, 13(4): 9735-9746. DOI:10.4238/2014.November.27.1
|
| [67] |
Zhang JW, Zhang CY, Wang JX, et al. Expression of the gene Lg-ATF1 encoding alcohol acetyltransferases from brewery lager yeast in Chinese rice wine yeast//Zhang TC, Ouyang P, Kaplan S, et al, eds. Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012). Berlin, Heidelberg: Springer, 2014, 249: 43-51.
|
| [68] |
Dong J, Xu HY, Zhao LB, et al. Enhanced acetate ester production of Chinese liquor yeast by overexpressing ATF1 through precise and seamless insertion of PGK1 promoter. J Ind Microbiol Biotechnol, 2014, 41(12): 1823-1828. DOI:10.1007/s10295-014-1522-4
|
| [69] |
Zhang CY, Liu YL, Qi YN, et al. Increased esters and decreased higher alcohols production by engineered brewer's yeast strains. Eur Food Res Technol, 2013, 236(6): 1009-1014. DOI:10.1007/s00217-013-1966-1
|
| [70] |
Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP, et al. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol, 2003, 69(9): 5228-5237. DOI:10.1128/AEM.69.9.5228-5237.2003
|
| [71] |
Li W, Wang JH, Zhang CY, et al. Regulation of Saccharomyces cerevisiae genetic engineering on the production of acetate esters and higher alcohols during Chinese Baijiu fermentation. J Ind Microbiol Biotechnol, 2017, 44(6): 949-960. DOI:10.1007/s10295-017-1907-2
|
| [72] |
Lian JZ, Si T, Nair NU, et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng, 2014, 24: 139-149. DOI:10.1016/j.ymben.2014.05.010
|
| [73] |
Park SH, Kim SJ, Hahn JS. Improvement of isobutanol production in Saccharomyces cerevisiae by increasing mitochondrial import of pyruvate through mitochondrial pyruvate carrier. Appl Microbiol Biotechnol, 2016, 100(17): 7591-7598. DOI:10.1007/s00253-016-7636-z
|
| [74] |
Feng RQ, Li JZ, Zhang AL. Improving isobutanol titers in Saccharomyces cerevisiae with over-expressing NADPH-specific glucose-6-phosphate dehydrogenase (Zwf1). Ann Microbiol, 2017, 67(12): 785-791. DOI:10.1007/s13213-017-1304-0
|
| [75] |
Wang ZY, Bai XJ, Guo XN, et al. Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol, 2017, 44(1): 129-139. DOI:10.1007/s10295-016-1852-5
|
| [76] |
Abe F, Horikoshi K. Enhanced production of isoamyl alcohol and isoamyl acetate by ubiquitination-deficient Saccharomyces cerevisiae mutants. Cell Mol Biol Lett, 2005, 10(3): 383-388.
|
| [77] |
孙中贯, 王孟祺, 王亚平, 等. GAP1 基因缺失对上面发酵酵母高级醇代谢能力的影响. 天津科技大学学报, 2020, 35(1): 10-17. Sun ZG, Wang MQ, Wang YP, et al. Effect of GAP1 gene deletion on higher alcohols production in top-fermenting yeast. J Tianjin Univ Sci Technol, 2020, 35(1): 10-17 (in Chinese). |
 2021, Vol. 37
2021, Vol. 37