中国科学院微生物研究所、中国微生物学会主办
文章信息
- 苏立秋, 张歌, 姚震, 梁配新, 戴宗杰, 王钦宏
- Su Liqiu, Zhang Ge, Yao Zhen, Liang Peixin, Dai Zongjie, Wang Qinhong
- 非传统酵母代谢工程研究进展
- Advances in metabolic engineering of non-conventional yeasts
- 生物工程学报, 2021, 37(5): 1659-1676
- Chinese Journal of Biotechnology, 2021, 37(5): 1659-1676
- 10.13345/j.cjb.200707
-
文章历史
- Received: November 3, 2020
- Accepted: February 14, 2021
- Published: March 8, 2021
2. 国家合成生物技术创新中心,天津 300308
2. National Technology Innovation Center of Synthetic Biology, Tianjin 300308, China
酵母在人类发展(游牧到农耕)、日常生活(面包、酒类饮品) 和科学研究(模式生物) 中扮演了不可或缺的角色,其中以酿酒酵母Saccharomyces cerevisiae和裂殖酵母Schizosaccharomyces pombe最为人熟知。这两种酵母被称为传统酵母(Conventional yeast),其他的酵母统称为非传统酵母(Non-conventional yeast)。
比较有代表性的非传统酵母有解脂耶氏酵母Yarrowia lipolytica、克鲁维酵母Kluyveromyces、毕赤酵母Pichia、假丝酵母Candida、汉逊酵母Hansenula等。与Crabtree阳性的酿酒酵母相比,这些酵母也被称为Crabtree阴性酵母,主要通过呼吸作用促进细胞生长,且具有许多优异的生理代谢性能[1]。其中,解脂耶氏酵母因乙酰-CoA和NADPH供应充足、蛋白糖基化水平低,主要用于生产蛋白、油脂、萜烯类、有机酸以及糖醇等[2];乳酸克鲁维酵母因具有代谢废乳清的能力,被广泛应用于食品和药物生产中蛋白质的合成[3];马克斯克鲁维酵母因具有广泛底物利用、快速生长、高温耐受等特点已成功用于生产乙醇及芳香化合物等[4];毕赤酵母和汉逊酵母具有高蛋白分泌能力和低糖基化水平,被广泛用于异源蛋白质的生产,同时因其天然的甲基营养型特点亦被用作一碳碳源利用底盘[5-6];假丝酵母因其具有高胁迫耐受性以及广底物利用谱也被大量研究和应用[7]。
随着系统生物学和合成生物学的发展,围绕这些非传统酵母的代谢工程研究取得了十分显著的进展。本文首先对上述非传统酵母中CRISPR-Cas基因编辑技术的进展进行归纳;然后从不同代谢工程改造策略角度总结了利用非传统酵母进行产品(非蛋白产品) 合成的研究进展;最后结合当前非传统酵母代谢工程改造过程中存在的问题对其未来可能的发展方向进行展望。
1 CRISPR-Cas基因编辑技术在非传统酵母中的应用进展高效精确的遗传改造工具和方法对于快速工程化重塑细胞代谢和实现目标产物的高效合成至关重要。目标DNA的编辑改造(基因敲除、基因整合) 都依赖于宿主细胞的DNA修复机制。在酿酒酵母中,同源重组(Homologous recombination,HR) 在双链断裂(Double-stranded break,DSB)修复过程中起主要作用[8],然而大多数非传统酵母主要利用非同源末端连接(Non-homologous end joining,NHEJ) 的方式进行DNA修复[9] (图 1),因此增强同源重组效率是实现在非传统酵母中基因精确改造和组装的必要条件。
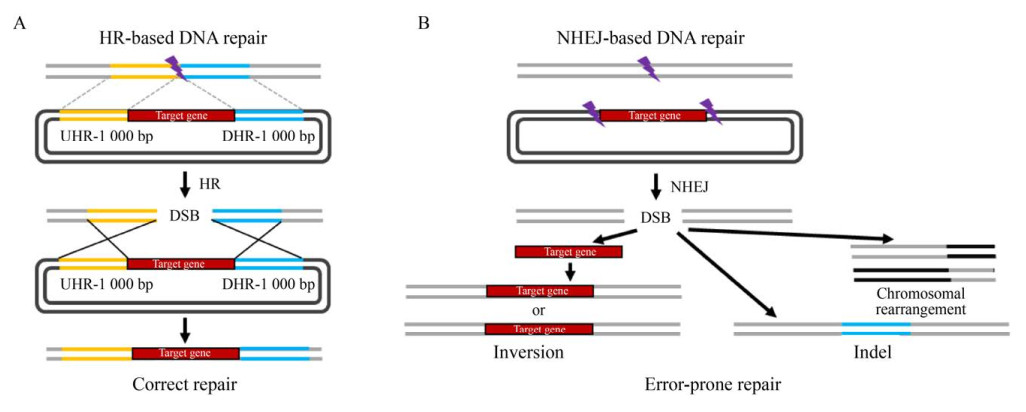
|
| 图 1 微生物细胞中主要的DNA修复方式 Fig. 1 Methods of DNA repair in microbial cells. |
| |
核酸内切酶Cas蛋白可以在基因组中精确高效地产生DSB,目前基于此发展的CRISPR-Cas基因编辑技术已被成功应用于解脂耶氏酵母、克鲁维酵母、毕赤酵母、假丝酵母、汉逊酵母等非传统酵母的遗传改造中,极大提高了非传统酵母的代谢工程改造进程[10]。以下将从表达gRNA启动子的选择和利用改造修饰Cas蛋白进行表达调控的两个角度对近些年CRISPR-Cas基因编辑技术在非传统酵母的研究进展进行综述。
gRNA的有效表达是利用CRISPR-Cas进行基因编辑的必要条件。在酿酒酵母中,依赖于Ⅲ型RNA聚合酶的Psnr52是最常用的gRNA表达启动子[11]。同样的,该启动子也可以在乳酸克鲁维酵母、白色假丝酵母和树干毕赤酵母中有效启动gRNA的表达[12-14]。在ku80敲除的乳酸克鲁维酵母中,基于Psnr52的CRISPR-Cas9系统成功地将具有1 kb同源臂的供体DNA整合到dit1、adh1和ndt80基因座,尽管三重整合效率仅为2%[12]。在白色假丝酵母中,Vyas等发现利用Psnr52基因组整合表达gRNA的基因编辑整合效率可达60%–80%[13]。在树干毕赤酵母中,利用该启动子的CRISPR-Cas9系统可以实现80%的插入缺失突变,在ku70/ku80双敲除菌株中,同源修复效率可达70%以上[14]。但在解脂耶氏酵母中利用snr52启动子表达gRNA的基因编辑效率偏低(13%),通过融合Ⅲ型RNA聚合酶启动子scr1与甘氨酸tRNA发现scr1-tRNAGly启动子启动表达gRNA的CRISPR-Cas9基因编辑系统基于HR修复的阳性率达64%,基于NHEJ修复的编辑效率达100%[15-16]。在马克斯克鲁维酵母中采用了类似的策略,通过融合Ⅲ型RNA聚合酶启动子rpr1与甘氨酸tRNA表达gRNA,可以实现XYL2的最高靶向效率达66%。利用该系统进行7个乙醇脱氢酶基因(adh1-7) 和atf的敲除,靶向效率为10%–67%[17]。在汉逊酵母中,采用融合snr6-tRNALeu启动子表达gRNA,将相关基因(pho1、pho11、pho84) 的插入缺失效率提高至17%–71%[18]。
依赖于Ⅱ型RNA聚合酶的启动子也被成功地用于非传统酵母中gRNA的表达,实现有效的基因编辑。Gao等在解脂耶氏酵母采用tefin启动子构建单一质粒来表达Cas9和多个gRNA,双基因敲除和三基因敲除的效率分别为36.7%和19.3%[19]。Weninger等在巴斯德毕赤酵母中利用Ⅱ型RNA聚合酶启动子Phxt1进行gRNA表达,基于NHEJ修复引入插入缺失突变的效率接近100%,在同源重组的片段上加入自主复制DNA序列,并将供体质粒线性化,在ku70敲除菌株中可以达到100%的同源重组效率[20]。在热带假丝酵母中利用Pfba1启动gRNA的表达,ura3单基因的同源修复率可达100%,利用Pgap1启动靶向ura3和ade2双gRNA的表达,在共同转化两个供体DNA的条件下,双基因的编辑效率为32%[21]。
除了利用CRISPR-Cas系统进行基因的插入缺失突变、基因敲除和基因整合表达外,近年来基于CRISPR-Cas系统的基因表达调控也在非传统酵母中得到了应用,主要是通过改造修饰相应Cas蛋白的表达来实现基因的表达调控。Yang等通过在巴斯德毕赤酵母中引入核酸内切酶失活的dCas9,将Aox1酶活水平降低了70%左右[22];类似地,Cao等利用dCas9将树干毕赤酵母中增强型绿色荧光蛋白(Enhanced green fluorescence protein,eGfp) 的表达量降低了17%[14];在马克斯克鲁维酵母中,采用dCas9的CRISPR技术同时下调4个基因(aco2b、sdh2、rip1、mss51) 的表达,将乙酸乙酯的生产强度提高了3.8倍[23];基于dCas9的CRISPR也被用于下调解脂耶氏酵母NHEJ相关基因的表达来提高HR的效率,其中下调ku70的表达可使HR效率达到56%,下调ku80后HR效率可达73%[24]。
通过融合dCas9和转录抑制因子Mix1可以进一步加强基因下调表达的强度,该技术被称为CRISPR干扰(CRISPR interference,CRISPRi)。利用该策略,树干毕赤酵母中eGfp的表达量相比没有融合Mix1的酵母可被进一步下调近1倍;相比未融合Mix1的酵母,融合表达dCas9和Mix1后解脂耶氏酵母Ku80的表达量也可被进一步下调约1.3倍[14]。另一方面通过融合表达dCas9与转录激活因子,可以实现目标基因的上调表达,该技术被称为CRISPR激活(CRISPR activation,CRISPRa)。融合表达dCaS9与转录激活结构域VPR激活β-葡萄糖苷酶的表达可以使解脂耶氏酵母在纤维二糖为唯一碳源的条件下生长[25]。另外通过控制gRNA的长度,表达未失活的Cas蛋白也可以实现目标基因的上调或下调。Ian研究团队通过截断表达16 nt的gRNA和融合表达Cas12a-Mix1、Cas12a-Vpr,可以将解脂耶氏酵母中can1的mRNA水平降低7倍,外源Gfp表达量提高10倍[26]。
2 非传统酵母代谢工程改造研究进展相比酿酒酵母,非传统酵母具有许多工业生产相关的生理代谢优势和特性,比如底物利用多样性、环境耐受性强、蛋白合成分泌能力强、生长速度快、适合高密度发酵等,这些特点吸引众多研究者通过各种代谢工程策略进一步提高非传统酵母的代谢性能,进行各类化学品的合成测试(表 1,图 2)。以下将从各类代谢工程改造策略的角度综述以非传统酵母为底盘的化学品合成的研究进展。

|
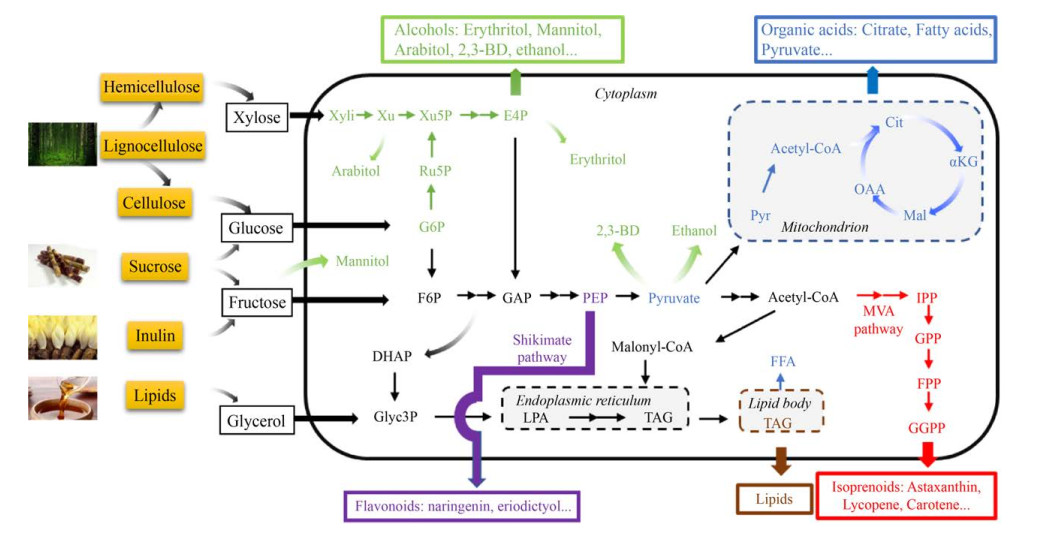
|
| 图 2 非传统酵母中底物利用和产物合成代谢途径概览 Fig. 2 Overview of pathways for substrate utilization and product synthesis in non-conventional yeast. |
| |
原料成本是决定生物合成化学品工业应用可行性的重要因素。为了降低生产成本,提高工业发酵可行性,研究者们基于非传统酵母的各自优势,利用代谢工程手段,拓宽生产菌株底物谱,使其可利用不同原料为碳源进行各类化学品的合成(图 2)。
解脂耶氏酵母因其降解疏水性碳源的能力而闻名。目前开发的解脂耶氏酵母疏水性碳源的来源包括纯油、食用废油和含油废水。解脂耶氏酵母UOFSY-1701菌株以葵花籽油为主要碳源生产柠檬酸,在添加乙酸盐的条件下,柠檬酸产量达到18.7 g/L[71]。由于食用废油的主要成分是甘油三酸酯,因此可以用作解脂耶氏酵母的原料,在10 L生物反应器中培养,解脂耶氏酵母SWJ-1b菌株从80 g/L食用废油中产生了31.7 g/L柠檬酸和6.5 g/L异柠檬酸[72]。含油废水中的脂质也被用作解脂耶氏酵母的原料,使用解脂耶氏酵母ACA-DC 50109菌株,将葡萄糖(65 g/L) 与含油废水结合使用可以产生效价为28.9 g/L的柠檬酸[73]。当含油废水与粗甘油混合时,柠檬酸产量和产率分别为37 g/L和0.55 g/g[74]。通过对脂耶氏酵母碳源的开发,促进了环境废弃资源的有效利用。
木糖是木质纤维素水解液中的主要成分,高效代谢木糖对于纤维素等低廉可再生原料的利用至关重要。非传统酵母利用木糖为碳源的研究非常广泛。在解脂耶氏酵母中通过表达来源于树干毕赤酵母的木糖还原酶和木糖醇脱氢酶以及内源木酮糖激酶,可以使解脂耶氏酵母以木糖为碳源生产柠檬酸,产量达80 g/L[75]。因为马克斯克鲁维酵母能够比大多数酵母在更高的温度下生长,所以其能在高温下有效地以木糖为碳源生产乙醇及木糖醇。通过在马克斯克鲁维酵母DMB1菌株中表达木糖还原酶,蛋白工程改造后的NADP+依赖的木糖醇脱氢酶及木酮糖激酶,可以利用葡萄糖和木糖在40 ℃条件下生产合成24.1 g/L乙醇,转化率达0.402 g/g[48]。在木糖醇生产中,马克斯克鲁维酵母YZJ074以木糖及甘油为碳源,在42 ℃生物反应器中产量可达312.05 g/L[76]。热带假丝酵母JA2以177 g/L木糖为碳源生产109.5 g/L木糖醇,生产效率和产率分别达2.8 g/(L·h) 和0.86 g/g[77]。在木糖醇脱氢酶基因敲除的热带假丝酵母中过表达来源于粗糙脉孢菌的木糖还原酶,以木糖为碳源,木糖醇的生产效率和产率分别为1.44 g/(L·h) 和96%,相比对照菌株分别提高73%和62%[78]。
甘油是生物柴油生产和工业皂化过程的主要副产品[79]。解脂耶氏酵母可利用甘油为天然碳源进行生长,过表达甘油激酶和甘油-3-磷酸脱氢酶,可使解脂耶氏酵母菌株A101的甘油利用率提高25%[80],柠檬酸产量仍可达34.1 g/L[81]。Gao等在解脂耶氏酵母菌株Po1f中敲除琥珀酸脱氢酶编码基因sdh5,并使用粗甘油为碳源,通过分批补料发酵,琥珀酸产量可达160 g/L[38]。Carly等通过在解脂耶氏酵母中过表达gut1 (甘油激酶编码基因) 和tkl1 (转酮酶编码基因),敲除eyk1 (赤藓酮糖激酶编码基因),使得以甘油为碳源的赤藓糖醇生产强度提高75%,发酵周期缩短40%[82]。在嗜热多形汉逊酵母中表达pdc1 (丙酮酸脱羧酶编码基因) 和adh1 (乙醇脱氢酶编码基因),在45 ℃发酵条件以甘油为碳源的乙醇产量为5.0 g/L,与野生型菌株相比提高5.4倍[83]。
糖蜜是制糖工业的主要副产品,其中含有大量可发酵糖(主要含有蔗糖、果糖等),因而是非常经济的发酵原料。解脂耶氏酵母由于缺乏蔗糖酶(Suc2) 而无法利用蔗糖。通过异源表达菊粉酶及来自酿酒酵母的suc2基因,解脂耶氏酵母的脂肪酸产量可达23.82 g/L,产率为0.16 g/g[28]。同样通过表达来自酿酒酵母的suc2基因,解脂耶氏酵母柠檬酸的产量和转化率可达45.02 g/L和0.643 g/g[84]。同时过表达己糖激酶(Hxk1) 及Suc2后,96 h发酵解脂耶氏酵母脂质含量达到9.15 g/L,生物量转化率为0.262 g/g,该菌株从甜菜糖蜜和甘油为碳源获得的最高脂质含量为0.31 g/g DCW,恒化发酵时脂质生产强度为0.43 g/(L·h)[85]。
菊粉是在不同植物中发现的果糖聚合物,已被用作生产乙醇和柠檬酸的廉价碳源。菊粉分解代谢的第一步是将其水解成可发酵的单糖。大多数非传统酵母无法自然代谢菊粉,据报道,在马克斯克鲁维酵母和木霉毕赤酵母中具有一定的菊粉酶活性。利用马克斯克鲁维酵母NRRL Y-50798自身菊粉酶活性,通过高温驯化,使其在46 ℃利用菊粉生产乙醇[86]。Liu等通过表达来源于马克斯克鲁维酵母CBS6556编码菊粉酶的inu1基因,获得了具有菊粉酶活性的解脂耶氏酵母重组菌株,在含有纯菊粉的生物反应器中柠檬酸浓度为68.9 g/L[87],通过重复分批培养,可使柠檬酸产量达203 g/L,产率及生产效率分别达0.85 g/g、0.51 g/(L·h)[37]。
2.2 增强乙酰辅酶A前体供应前体物质供给充足是保证目标产品高产的必要条件。在众多代谢物中,乙酰辅酶A是生物体合成代谢和分解代谢的中心枢纽,也是众多化学品合成的必需前体。因此为了提高目标产物合成,研究者们围绕非传统酵母中乙酰辅酶A的供给进行了大量研究(图 3)。

|
| 图 3 解脂耶氏酵母中胞质乙酰-CoA调控常用策略 Fig. 3 Common strategies for the regulation of cytoplasmic acetyl-CoA in Yarrowia lipolytica. |
| |
解脂耶氏酵母利用ATP柠檬酸裂解酶(Acl)提供乙酰辅酶A。为了提高胞质乙酰辅酶A的合成,将小家鼠Mus musculus来源的acl基因导入多拷贝整合型质粒并在解脂耶氏酵母中进行表达,使脂质含量由7.3%提高到23%[88]。Xu等在解脂耶氏酵母中分别设计并比较了5种不同胞质乙酰辅酶A替代途径[89],发现这些途径在过表达乙酰辅酶A羧化酶和二酰基甘油转移酶的工程菌株中表达时,均能够提高脂质的效价和含油量,其中肉碱乙酰转移酶Cat2过表达菌株的脂质产量可达66.4 g/L,相比对照提高约3倍。另一研究中,在改变菌株的底物利用谱后,基于构建的木糖利用途径,即过表达磷酸酮醇酶(XpkA)、乙酸激酶(Ack) 和乙酰辅酶A合成酶(Acs),进一步增强了乙酰CoA供给;在木质纤维素水解液为原料的分批补料生物反应器中,脂质产量为16.5 g/L,与野生菌株相比提高8.3倍[27]。在脂肪酸乙酯(Fatty acid ethyl esters,FAEE) 生产中,Gao等通过过表达自身柠檬酸裂解酶编码基因acl1、acl2,酿酒酵母的乙酰辅酶A合成酶基因acs2,使FAEE的产量提高40%[35]。
乙酰辅酶A的供给在有机酸及醇类物质生产中也发挥重要作用。丙酮酸是发酵生产α-酮戊二酸过程中主要副产物。Zhou等在高产α-酮戊二酸的解脂耶氏酵母菌株WSH-Z06中,分别过表达了来自酿酒酵母的Acs和来源于小鼠的ATP柠檬酸裂合酶Acl1,以提高乙酰辅酶A的转化率,降低丙酮酸积累,所获重组菌株的α-酮戊二酸产量均有提高,发酵过表达acl基因的重组菌株,α-酮戊二酸产量可达56.5 g/L,丙酮酸产量从35.1 g/L降低至20.2 g/L[90]。在产朊假丝酵母生产异丙醇过程中,为了增加乙酰辅酶A的供应,Tamakawa等着重研究了与乙酰辅酶A产生有关的乙醇同化途径(乙醇通过乙醇脱氢酶(Adh1、Adh2),乙醛脱氢酶(Ald6) 和乙酰辅酶A合酶(Acs1、Acs2) 的作用转化为乙酰辅酶A) 和乙酰辅酶A乙酰转移酶(Erg10) 对异丙醇合成的影响。通过表达acs1、acs2及erg10基因分别使异丙醇产量提高2.5倍、2.7倍及2.2倍。通过将三拷贝acs2与六拷贝erg10在宿主菌中共表达,与原始菌株相比,产量提高11.9倍,经分批补料发酵,异丙醇产量达到27.2 g/L[51]。
2.3 强化目标产物合成途径代谢通量增强产物合成途径中酶的表达,减少分支代谢途径的竞争是提高目标产物合成效率的基本代谢工程策略。该策略被广泛应用于各类底盘微生物高效合成众多产品的代谢工程改造中,其中萜烯类产品的微生物合成就是典型示例。
萜烯类物质是一类重要的天然香料和前体物质,是化妆品、食品和工业制造的关键原料,同时也发现许多萜烯化合物为中草药的有效组成成分[91]。在酵母中萜烯类化合物的前体物质异戊烯基二磷酸酯(Isopentenyl diphosphate ester,IPP)和二甲基烯丙基二磷酸酯(Dimethylallyl diphosphate ester,DMAPP) 主要通过甲羟戊酸(Mevalonic acid,MVA) 途径生成,进一步通过异戊二烯基二磷酸合酶(Idi1) 等合酶作用生成单萜前体香叶基二磷酸(Geraniyl diphosphate,GPP,C10)、倍半萜前体法呢基二磷酸(Farnesyl diphosphate,FPP,C15)、二萜前体香叶基香叶基二磷酸(Geraniyl geraniyl diphosphate,GGPP,C20),两个FPP或GGPP分子缩合生成三萜烯(C30) 或四萜烯(C40) 合成前体。所得的链状类异戊二烯物质在萜类合酶催化下进一步修饰,从而产生大量结构复杂的天然产物(图 4)。以下将以萜烯类产品的合成为代表,从内源关键基因的表达与调控、旁路途径的弱化、外源关键酶的筛选与表达3个方面来阐述非传统酵母萜类化合物的合成进展。

|
| 图 4 非传统酵母中萜类化合物合成途径 Fig. 4 Terpenoids synthesis pathway in non-conventional yeast. |
| |
HMG-CoA还原酶(HmgR)、异戊二烯基二磷酸合酶(Idi1) 被认为是甲羟戊酸途径关键限速酶。截短HmgR (tHmgR) 的N末端氨基酸可以使其在解脂耶氏酵母细胞质中稳定存在。Idi1催化IPP生成DMAPP,此酶的过表达可以平衡IPP和DMAPP含量,在GPP和FPP的分布中起重要作用。tHmgR及Idi1的过表达是加强萜类物质合成的常用策略。过表达tHmgR使得解脂酵母中芳樟醇的产量提高了4.7倍,在此基础上对Idi1进行表达强化使芳樟醇产量提高2.8倍[56]。Yang等通过在解脂酵母中过表达tHmgR编码基因thmg1,使法尼烯产量提高12%,同时比较了在含有及不含有thmg1过表达情况下,idi1过表达对产量的影响,发现当thmg1过表达时,idi1过表达使法尼烯产量提高两倍[92]。此外,MVA代谢途径其他关键酶Erg8、Erg10、Erg12和Erg19也被认为可以增强萜类化合物的前体供应。Liu等通过将erg8、erg12、erg19、idi1基因在解脂酵母中进行过表达,最终使法尼烯产量提高72%[57]。
2.3.2 旁路途径的弱化Erg20既是GPP合酶,又是FPP合酶,该双功能特性导致以GPP为前体的单萜合成效率较低。Ignea等报道,Erg20的突变(即F96W和N127W) 可以使酿酒酵母中的碳通量从FPP转移到GPP,显著影响酿酒酵母单萜物质的生产[93]。在解脂耶氏酵母生产芳樟醇过程中,为了确保GPP的足够可用性,Cao等采用了类似的策略来修饰解脂耶氏酵母中的Erg20,基于与酿酒酵母Erg20的氨基酸序列比对分析,确定了解脂耶氏酵母Erg20的氨基残基为F88和N119,向解脂耶氏酵母菌株CXY01中引入突变体erg20F88W-N119W,所得菌株CXY36的芳樟醇产量为5.34 mg/L,比亲本菌株高3.7倍[56]。
FPP是细胞自身组分(包括泛醌、角鲨烯、胆固醇、GGPP和其他必需固醇) 的重要前体。角鲨烯合酶Sqs1催化两个FPP分子缩合为角鲨烯,完全抑制Sqs1活性会严重影响细胞生长,因此将sqs1表达下调是促进萜类生产的常用策略。Kildegaard等通过截短sqs1启动子,或者将其启动子替换为Perg1或Perg11,下调解脂耶氏酵母中类胡萝卜素高产菌株的角鲨烯通量,与带有自身sqs1启动子的对照菌株相比,所得菌株的β-胡萝卜素产量提高了2.0–2.5倍,其中sqs1启动子截短到50 bp菌株的β-胡萝卜素产量接近800 mg/L[62]。在类胡萝卜素相关物质生产中,除了代谢下游途径需要弱化,干扰负责胞内脂质代谢的β氧化途径也可以促进解脂酵母中番茄红素的合成。Matthäus等在解脂耶氏酵母中将β氧化途径的pox1-6基因及甘油三磷酸脱氢酶gut2基因敲除,阻断脂肪酸β氧化,减少甘油三酯的形成,从而促进了脂质体的生成,通过补料发酵每克细胞干重番茄红素产量可以达到16 mg[94]。
2.3.3 外源关键酶的筛选与表达在天然产物合成中,需要外源引入将MVA途径链状化合物转化为多环骨架的萜烯合酶及对萜烯合酶进行氧化修饰的细胞色素P450酶。萜烯合酶及P450酶的高活性对萜类物质的生物合成十分重要。圆柚酮属于倍半萜类物质,广泛用于香料和化妆品行业。通过将来源于黄金柏的朱栾倍半萜合酶CnVS、P450酶Cyp706M1和来源于拟南芥的P450还原酶AtCpr1共表达于解脂耶氏酵母中,可使解脂耶氏酵母获得圆柚酮生产能力。为了提高圆柚酮产量,进一步将cyp706m1及Atcpr1融合表达,促进P450酶之间的电子传递,最终使产量提高6倍[64]。同样在毕赤酵母中异源表达钝莨菪的加氧酶和拟南芥的细胞色素P450还原酶,可以有效地将外源添加的朱栾倍半萜合成圆柚酮,通过外源表达朱栾倍半萜合酶,可以解除对朱栾倍半萜的依赖。通过过表达醇脱氢酶和tHmg1,增加前体供应,再通过两相发酵,最终将圆柚酮的产量提高到208 mg/L。如P450酶及P450还原酶融合表达的蛋白工程手段已在非传统酵母中广泛应用,尤其将外源萜类合酶与内源GPP/FPP合酶的融合表达[65]。Yang等通过将来源于苹果的法尼烯合酶在解脂耶氏酵母中表达,使菌株具有合成法尼烯的能力,为了促进FPP向法尼烯的转化,将FPP合酶及法尼烯合酶进行融合表达,使法尼烯的产量与单独过表达法尼烯合酶相比提高4倍[92]。
2.4 细胞生理代谢的全局优化 2.4.1 辅因子调控平衡产物合成和细胞生理代谢所需的氧化还原状态是实现代谢流高效导向目标代谢产物的必要手段。由于氧受限条件易引起细胞氧化还原失衡,酵母将木糖高效转化为乙醇存在一定的挑战。例如:耐热马克斯克鲁维酵母可以以木糖为碳源在高温下生长良好,但其木糖发酵能力较弱。Zhang等通过外源引入来自粗糙脉孢菌的NADPH依赖的木糖还原酶(Xr) 和来自树干毕赤酵母的NADP+依赖的木糖醇脱氢酶(Xdh) 突变体,结合下游基因的过表达,最终使重组菌株的木糖发酵能力和氧化还原平衡均得到了显著提高。所得工程菌株YZJ088在42 ℃条件下利用118.4 g/L木糖产生44.9 g/L乙醇,生产强度达2.49 g/(L·h)[49]。
NADPH供给是脂肪酸和脂质合成中的主要限速因素。在脂质生产中,为了高效利用物质代谢还原力以提高脂质产量,Qiao等在解脂耶氏酵母中将糖酵解途径产生的NADH转化为脂质生物合成需要的NADPH合成途径。在过表达acc1和dga1的菌株中进一步过表达丙酮丁醇梭菌来源的gapC和卷枝毛霉来源的mce2,使胞内NADH有效转化为NADPH,最终使工程菌株的产率达0.231 g/g,与对照相比提高了25%[29]。为了循环使用胞质中NADPH提高解脂耶氏酵母三乙酸内酯(Triacetate lactone,TAL) 的合成,Liu等测试了一系列胞质NADPH合成途径(苹果酸酶、甘露醇脱氢酶、6-磷酸葡萄糖酸脱氢酶、琥珀酸半醛脱氢酶) 是否会改善TAL的合成和细胞适应性。其中内源NADP+依赖型苹果酸酶(Mae) 的过表达可显著提高TAL产量。当ylMae与TAL合成关键酶Gh2ps及ylAcc1共同过表达时,使菌株TAL增产60%[95]。NADPH的循环利用还可有效促进萜类物质的生产。HMG-CoA还原酶(HmgR) 是甲羟戊酸途径中的第一个限速酶,在调节角鲨烯的生物合成中起关键作用,HMG-CoA通过以NADPH作为还原力释放辅酶A后被还原水解为甲羟戊酸。Liu等通过将甘露醇脱氢酶(通过甘露醇循环调控胞质NADHP含量) 引入解脂耶氏酵母代谢系统,使鲨烯产量提高11%[96]。在三萜类物质生产中,NADPH是内源甲羟戊酸途径和异源萜类合酶P450氧化还原系统的直接氧化还原供应物质。Jin等通过在解脂耶氏酵母中过表达苹果酸脱氢酶编码基因(emt及rtme) 使胞内NADPH供给提高,最终促进了三萜类物质桦木酸的生产[66]。
2.4.2 转录因子调控转录因子作为信号分子参与细胞生理代谢调控的众多方面。通过改变控制特定代谢途径转录因子的表达,可以避免对合成途径相关酶的表达进行个体调谐,从而在高层级上对目标产物合成途径进行全局优化,有效提高目标途径代谢通量及产物合成效率。研究表明来自汉逊德巴利酵母的转录因子Sef1对核黄素生物合成途径的表达进行正向调节。Dmytruk等在假丝酵母突变株AF-4中通过引入来自汉逊德巴利酵母的sef1,使核黄素产量提高87.2%[97]。在解脂耶氏酵母中,研究表明参与葡萄糖阻遏作用的转录因子Snf1及Mig1对胞内脂质合成具有调节作用。Snf1参与解脂耶氏酵母由生长阶段向产脂阶段的转变,该转录因子的敲除使得菌体脂肪酸产量提高2.6倍,经转录分析发现Snf1敲除菌中脂质合成相关基因明显上调[98]。类似的研究发现敲除Mig1会导致解脂耶氏酵母脂肪酸β-氧化途径mfe1的转录下调和与脂质合成相关的基因上调,敲除该基因的菌株胞内油脂含量提高到细胞干重的48.7%,相比对照菌株提高1.3倍[99]。
2.4.3 适应性进化胞内代谢途径的加强和胞外环境的胁迫都会增加细胞的代谢负担,适应性进化是提高并稳定菌株生产性状的重要策略。这种策略目前已在非传统酵母的碳源利用及环境耐受的研究中广泛应用。Sharma等在45 ℃以木糖为碳源的条件下,对耐热酵母马克斯克鲁维酵母进行多达60批次的适应进化,以提高其木糖利用能力。与天然菌株相比,进化菌株显示出更高的比生长速率和更快的木糖摄取速率以及更短的生长延滞期。厌氧发酵时,进化菌株在72 h内消耗约91%的木糖,分别产生了2.88 g/L和18.75 g/L的乙醇和木糖醇,相比未进化菌株提高约5倍,乙醇和木糖醇的得率也提高约1.5倍[100]。在高浓度葡萄糖环境中,对可以从葡萄糖合成D-阿拉伯醇的巴斯德毕赤酵母GS115菌株进行驯化,得到驯化菌株GS225,其D-阿拉伯醇的产量相比未驯化菌株GS115提高近一倍,达到12.5 g/L[101]。同样通过对可利用CO2的巴斯德毕赤酵母适应性进化,使其以CO2为唯一碳源时比生长速率由0.008 /h提高到0.018/h[102]。Daskalaki等采用适应性实验室进化策略对解脂耶氏酵母进行驯化,通过交替改变培养基成分并连续筛选高脂细胞,最终将细胞生长与脂质积累相偶联。通过77代筛选后,菌株的脂质含量达到细胞干重的44%,比原始菌株提高30%[103]。
3 总结与展望由于具有优良的天然特性,解脂耶氏酵母、克鲁维酵母、毕赤酵母、假丝酵母等非传统酵母在生物技术开发及工业应用中起着举足轻重的作用。随着高效基因操作工具的开发及代谢途径调控策略的完善使用,将非传统酵母改造为脂质、天然产物、有机酸及醇类物质高效合成细胞工厂已成为可能。但目前大多数非传统酵母菌的目标物质产量较低,远未使其优良天然特性得到充分发挥,限制了其在工业生产中的应用。多种因素影响其生产性能(产量、产率、转化率) 的改善,包括高效基因编辑工具开发、胞内代谢网络的高清解析、胞外环境互作的应答机制等。
目前在产品生产性能优化中主要集中于有利表型直接代谢途径的优化改造,缺乏关于代谢网络相互作用和代谢平衡调控与应答的关键基础信息,阻碍了非传统酵母在产品应用中的发展。同时高效关键酶的挖掘及改造对产物生成起关键作用,对非传统酵母的改造中针对目标途径优良酶的筛选主要基于文献报道或基因库中筛选,而基于关键酶进行定向进化及理性设计以提高关键酶催化活性的策略应用较少,阻碍了关键途径催化效率的最大化提升。这些领域的基本信息不全是建造产业化应用水平非传统酵母细胞工厂的主要障碍。相信多组学整合技术、高效基因编辑技术、多信号输入输出的动态调控技术以及高通量筛选技术等系统生物学与合成生物学的进步与发展,将促进人们对非传统酵母的独特生理代谢特征的深入了解,在最大化改造利用非传统酵母优良表型、提高产品生产性能、达到规模化工业应用水平等方面将具有重大突破。
| [1] |
Wagner JM, Alper HS. Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet Biol, 2016, 89: 126-136. DOI:10.1016/j.fgb.2015.12.001
|
| [2] |
Ma JB, Gu Y, Marsafari M, et al. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J Ind Microbiol Biotechnol, 2020, 47(9/10): 845-862. DOI:10.1007/s10295-020-02290-8
|
| [3] |
Spohner SC, Schaum V, Quitmann H, et al. Kluyveromyces lactis: an emerging tool in biotechnology. J Biotechnol, 2016, 222: 104-116. DOI:10.1016/j.jbiotec.2016.02.023
|
| [4] |
Löbs AK, Lin JL, Cook M, et al. High throughput, colorimetric screening of microbial ester biosynthesis reveals high ethyl acetate production from Kluyveromyces marxianus on C5, C6, and C12 carbon sources. Biotechnol J, 2016, 11(10): 1274-1281. DOI:10.1002/biot.201600060
|
| [5] |
Van Der Klei IJ, Yurimoto H, Sakai Y, et al. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta, 2006, 1763(12): 1453-1462. DOI:10.1016/j.bbamcr.2006.07.016
|
| [6] |
Nocon J, Steiger MG, Pfeffer M, et al. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng, 2014, 24: 129-138. DOI:10.1016/j.ymben.2014.05.011
|
| [7] |
Ju JH, Oh BR, Heo SY, et al. Production of adipic acid by short- and long-chain fatty acid acyl-CoA oxidase engineered in yeast Candida tropicalis. Bioprocess Biosyst Eng, 2020, 43(1): 33-43. DOI:10.1007/s00449-019-02202-w
|
| [8] |
Fraczek MG, Naseeb S, Delneri D. History of genome editing in yeast. Yeast, 2018, 35(5): 361-368. DOI:10.1002/yea.3308
|
| [9] |
Löbs AK, Schwartz C, Wheeldon I. Genome and metabolic engineering in non-conventional yeasts: current advances and applications. Synth Syst Biotechnol, 2017, 2(3): 198-207. DOI:10.1016/j.synbio.2017.08.002
|
| [10] |
Raschmanová H, Weninger A, Glieder A, et al. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol Adv, 2018, 36(3): 641-665. DOI:10.1016/j.biotechadv.2018.01.006
|
| [11] |
DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res, 2013, 41(7): 4336-4343. DOI:10.1093/nar/gkt135
|
| [12] |
Horwitz AA, Walter JM, Schubert MG, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst, 2015, 1(1): 88-96. DOI:10.1016/j.cels.2015.02.001
|
| [13] |
Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv, 2015, 1(3): e1500248. DOI:10.1126/sciadv.1500248
|
| [14] |
Cao MF, Gao MR, Ploessl D, et al. CRISPR-mediated genome editing and gene repression in Scheffersomyces stipitis. Biotechnol J, 2018, 13(9): 1700598. DOI:10.1002/biot.201700598
|
| [15] |
Schwartz CM, Hussain MS, Blenner M, et al. Synthetic RNA polymerase Ⅲ promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol, 2016, 5(4): 356-359. DOI:10.1021/acssynbio.5b00162
|
| [16] |
Schwartz C, Shabbir-Hussain M, Frogue K, et al. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synth Biol, 2017, 6(3): 402-409. DOI:10.1021/acssynbio.6b00285
|
| [17] |
Löbs AK, Engel R, Schwartz C, et al. CRISPR-Cas9-enabled genetic disruptions for understanding ethanol and ethyl acetate biosynthesis in Kluyveromyces marxianus. Biotechnol Biofuels, 2017, 10: 164. DOI:10.1186/s13068-017-0854-5
|
| [18] |
Numamoto M, Maekawa H, Kaneko Y. Efficient genome editing by CRISPR/Cas9 with a tRNA-sgRNA fusion in the methylotrophic yeast Ogataea polymorpha. J Biosci Bioeng, 2017, 124(5): 487-492. DOI:10.1016/j.jbiosc.2017.06.001
|
| [19] |
Gao SL, Tong YY, Wen ZQ, et al. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J Ind Microbiol Biotechnol, 2016, 43(8): 1085-1093. DOI:10.1007/s10295-016-1789-8
|
| [20] |
Cao MF, Gao MR, Lopez-Garcia CL, et al. Centromeric DNA facilitates nonconventional yeast genetic engineering. ACS Synth Biol, 2017, 6(8): 1545-1553. DOI:10.1021/acssynbio.7b00046
|
| [21] |
Zhang LH, Zhang HB, Liu YF, et al. A CRISPR-Cas9 system for multiple genome editing and pathway assembly in Candida tropicalis. Biotechnol Bioeng, 2020, 117(2): 531-542. DOI:10.1002/bit.27207
|
| [22] |
Yang YK, Liu GQ, Chen X, et al. High efficiency CRISPR/Cas9 genome editing system with an eliminable episomal sgRNA plasmid in Pichia pastoris. Enzyme Microb Technol, 2020, 138: 109556. DOI:10.1016/j.enzmictec.2020.109556
|
| [23] |
Löbs AK, Schwartz C, Thorwall S, et al. Highly multiplexed CRISPRi repression of respiratory functions enhances mitochondrial localized ethyl acetate biosynthesis in Kluyveromyces marxianus. ACS Synth Biol, 2018, 7(11): 2647-2655. DOI:10.1021/acssynbio.8b00331
|
| [24] |
Schwartz C, Frogue K, Ramesh A, et al. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnol Bioeng, 2017, 114(12): 2896-2906. DOI:10.1002/bit.26404
|
| [25] |
Schwartz C, Curtis N, Löbs AK, et al. Multiplexed CRISPR activation of cryptic sugar metabolism enables Yarrowia Lipolytica growth on cellobiose. Biotechnol J, 2018, 13(9): 1700584. DOI:10.1002/biot.201700584
|
| [26] |
Yang ZJ, Edwards H, Xu P. CRISPR-Cas12a/ Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab Eng Commun, 2019, 10: e00112.
|
| [27] |
Niehus X, Coq AMCL, Sandoval G, et al. Engineering Yarrowia lipolytica to enhance lipid production from lignocellulosic materials. Biotechnol Biofuels, 2018, 11: 11. DOI:10.1186/s13068-018-1010-6
|
| [28] |
Hapeta P, Rakicka M, Dulermo R, et al. Transforming sugars into fat-lipid biosynthesis using different sugars in Yarrowia lipolytica. Yeast, 2017, 34(7): 293-304. DOI:10.1002/yea.3232
|
| [29] |
Qiao KJ, Wasylenko TM, Zhou K, et al. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol, 2017, 35(2): 173-177. DOI:10.1038/nbt.3763
|
| [30] |
Yan FX, Dong GR, Qiang S, et al. Overexpression of Δ12, Δ15-desaturases for enhanced lipids synthesis in Yarrowia lipolytica. Front Microbiol, 2020, 11: 289. DOI:10.3389/fmicb.2020.00289
|
| [31] |
Yuzbasheva EY, Mostova EB, Andreeva NI, et al. A metabolic engineering strategy for producing free fatty acids by the Yarrowia lipolytica yeast based on impairment of glycerol metabolism. Biotechnol Bioeng, 2018, 115(2): 433-443. DOI:10.1002/bit.26402
|
| [32] |
Hanko EKR, Denby CM, I Nogué VS, et al. Engineering β-oxidation in Yarrowia lipolytica for methyl ketone production. Metab Eng, 2018, 48: 52-62. DOI:10.1016/j.ymben.2018.05.018
|
| [33] |
Bruder S, Moldenhauer EJ, Lemke RD, et al. Drop-in biofuel production using fatty acid photodecarboxylase from Chlorella variabilis in the oleaginous yeast Yarrowia lipolytica. Biotechnol Biofuels, 2019, 12: 202. DOI:10.1186/s13068-019-1542-4
|
| [34] |
Cordova LT, Butler J, Alper HS. Direct production of fatty alcohols from glucose using engineered strains of Yarrowia lipolytica. Metab Eng Commun, 2020, 10: e00105. DOI:10.1016/j.mec.2019.e00105
|
| [35] |
Gao Q, Cao X, Huang YY, et al. Overproduction of fatty acid ethyl esters by the oleaginous yeast Yarrowia lipolytica through metabolic engineering and process optimization. ACS Synth Biol, 2018, 7(5): 1371-1380. DOI:10.1021/acssynbio.7b00453
|
| [36] |
Chattopadhyay A, Gupta A, Maiti MK. Engineering an oleaginous yeast Candida tropicalis SY005 for enhanced lipid production. Appl Microbiol Biotechnol, 2020, 4(19): 8399-8411. DOI:10.1007/s00253-020-10830-6
|
| [37] |
Rakicka M, Wolniak J, Lazar Z, et al. Production of high titer of citric acid from inulin. BMC Biotechnol, 2019, 19: 11. DOI:10.1186/s12896-019-0503-0
|
| [38] |
Gao CJ, Yang XF, Wang HM, et al. Robust succinic acid production from crude glycerol using engineered Yarrowia lipolytica. Biotechnol Biofuels, 2016, 9(1): 179. DOI:10.1186/s13068-016-0597-8
|
| [39] |
Zhao C, Cui ZY, Zhao XY, et al. Enhanced itaconic acid production in Yarrowia lipolytica via heterologous expression of a mitochondrial transporter MTT. Appl Microbiol Biotechnol, 2019, 3(5): 2181-2192.
|
| [40] |
Sun W, Vila-Santa A, Liu N, et al. Metabolic engineering of an acid-tolerant yeast strain Pichia kudriavzevii for itaconic acid production. Metab Eng Commun, 2020, 10: e00124. DOI:10.1016/j.mec.2020.e00124
|
| [41] |
Chen XL, Li Y, Tong T, et al. Spatial modulation and cofactor engineering of key pathway enzymes for fumarate production in Candida glabrata. Biotechnol Bioeng, 2019, 116(3): 622-630. DOI:10.1002/bit.26906
|
| [42] |
Guo F, Dai Z, Peng W, et al. Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol Bioeng, 2020, 118(1): 357-371.
|
| [43] |
Zhu MY, Sun L, Lu XY, et al. Establishment of a transient CRISPR-Cas9 genome editing system in Candida glycerinogenes for co-production of ethanol and xylonic acid. J Biosci Bioeng, 2019, 128(3): 283-289. DOI:10.1016/j.jbiosc.2019.03.009
|
| [44] |
Wang JQ, Peng J, Fan H, et al. Development of mazF-based markerless genome editing system and metabolic pathway engineering in Candida tropicalis for producing long-chain dicarboxylic acids. J Ind Microbiol Biotechnol, 2018, 45(11): 971-981. DOI:10.1007/s10295-018-2074-9
|
| [45] |
Cheng HL, Wang SQ, Bilal M, et al. Identification, characterization of two NADPH-dependent erythrose reductases in the yeast Yarrowia lipolytica and improvement of erythritol productivity using metabolic engineering. Microb Cell Fact, 2018, 17: 133. DOI:10.1186/s12934-018-0982-z
|
| [46] |
Chi P, Wang SQ, Ge XM, et al. Efficient D-threitol production by an engineered strain of Yarrowia lipolytica overexpressing xylitol dehydrogenase gene from Scheffersomyces stipitis. Biochem Eng J, 2019, 149: 107259. DOI:10.1016/j.bej.2019.107259
|
| [47] |
Hua Y, Wang JC, Zhu YL, et al. Release of glucose repression on xylose utilization in Kluyveromyces marxianus to enhance glucose-xylose co-utilization and xylitol production from corncob hydrolysate. Microb Cell Fact, 2019, 18: 24. DOI:10.1186/s12934-019-1068-2
|
| [48] |
Suzuki T, Hoshino T, Matsushika A. High-temperature ethanol production by a series of recombinant xylose-fermenting Kluyveromyces marxianus strains. Enzyme Microb Technol, 2019, 129: 109359. DOI:10.1016/j.enzmictec.2019.109359
|
| [49] |
Zhang J, Zhang B, Wang DM, et al. Rapid ethanol production at elevated temperatures by engineered thermotolerant Kluyveromyces marxianus via the NADP(H)-preferring xylose reductase-xylitol dehydrogenase pathway. Metab Eng, 2015, 31: 140-152. DOI:10.1016/j.ymben.2015.07.008
|
| [50] |
Zong H, Zhang C, Zhuge B, et al. Effects of xylitol dehydrogenase (XYL2) on xylose fermentation by engineered Candida glycerinogenes. Biotechnol Appl Biochem, 2017, 64(4): 590-599. DOI:10.1002/bab.1514
|
| [51] |
Tamakawa H, Mita T, Yokoyama A, et al. Metabolic engineering of Candida utilis for isopropanol production. Appl Microbiol Biotechnol, 2013, 97(14): 6231-6239. DOI:10.1007/s00253-013-4964-0
|
| [52] |
Siripong W, Wolf P, Kusumoputri TP, et al. Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate. Biotechnol Biofuels, 2018, 11: 1. DOI:10.1186/s13068-017-1003-x
|
| [53] |
Yang ZL, Zhang ZS. Production of (2R, 3R)-2, 3- butanediol using engineered Pichia pastoris: strain construction, characterization and fermentation. Biotechnol Biofuels, 2018, 11: 35. DOI:10.1186/s13068-018-1031-1
|
| [54] |
Kong SJ, Pan H, Liu XY, et al. De novo biosynthesis of 2-phenylethanol in engineered Pichia pastoris. Enzyme Microb Technol, 2020, 133: 109459. DOI:10.1016/j.enzmictec.2019.109459
|
| [55] |
Cheng BQ, Wei LJ, Lv YB, et al. Elevating limonene production in oleaginous yeast Yarrowia lipolytica via genetic engineering of limonene biosynthesis pathway and optimization of medium composition. Biotechnol Bioproc Eng, 2019, 24(3): 500-506. DOI:10.1007/s12257-018-0497-9
|
| [56] |
Cao X, Wei LJ, Lin JY, et al. Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour Technol, 2017, 245: 1641-1644. DOI:10.1016/j.biortech.2017.06.105
|
| [57] |
Liu YH, Jiang X, Cui ZY, et al. Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene. Biotechnol Biofuels, 2019, 12: 296. DOI:10.1186/s13068-019-1636-z
|
| [58] |
Zhang XY, Wang DG, Duan YH, et al. Production of lycopene by metabolically engineered Pichia pastoris. Biosci Biotechnol Biochem, 2020, 84(3): 463-470. DOI:10.1080/09168451.2019.1693250
|
| [59] |
Gao SL, Tong YY, Zhu L, et al. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous β-carotene production. Metab Eng, 2017, 41: 192-201. DOI:10.1016/j.ymben.2017.04.004
|
| [60] |
Araya-Garay JM, Feijoo-Siota L, Rosa-dos-Santos F, et al. Construction of new Pichia pastoris X-33 strains for production of lycopene and β-carotene. Appl Microbiol Biotechnol, 2012, 93(6): 2483-2492. DOI:10.1007/s00253-011-3764-7
|
| [61] |
Tramontin LRR, Kildegaard KR, Sudarsan S, et al. Enhancement of astaxanthin biosynthesis in oleaginous yeast Yarrowia lipolytica via microalgal pathway. Microorganisms, 2019, 7(10): 472. DOI:10.3390/microorganisms7100472
|
| [62] |
Kildegaard KR, Adiego-Pérez B, Belda DD, et al. Engineering of Yarrowia lipolytica for production of astaxanthin. Synth Syst Biotechnol, 2017, 2(4): 287-294. DOI:10.1016/j.synbio.2017.10.002
|
| [63] |
Araya-Garay JM, Ageitos JM, Vallejo JA, et al. Construction of a novel Pichia pastoris strain for production of xanthophylls. AMB Expr, 2012, 2: 24. DOI:10.1186/2191-0855-2-24
|
| [64] |
Guo XY, Sun J, Li DS, et al. Heterologous biosynthesis of (+)-nootkatone in unconventional yeast Yarrowia lipolytica. Biochem Eng J, 2018, 137: 125-131. DOI:10.1016/j.bej.2018.05.023
|
| [65] |
Wriessnegger T, Augustin P, Engleder M, et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng, 2014, 24: 18-29. DOI:10.1016/j.ymben.2014.04.001
|
| [66] |
Jin CC, Zhang JL, Song H, et al. Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microb Cell Fact, 2019, 18: 77. DOI:10.1186/s12934-019-1127-8
|
| [67] |
Lu YP, Yang QY, Lin ZL, et al. A modular pathway engineering strategy for the high-level production of β-ionone in Yarrowia lipolytica.Microb. Cell Fact,, 2020, 19: 49. DOI:10.1186/s12934-020-01309-0
|
| [68] |
Marsafari M, Xu P. Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica. Metab Eng Commun, 2020, 10: e00121. DOI:10.1016/j.mec.2019.e00121
|
| [69] |
Wu YF, Xu S, Gao X, et al. Enhanced protopanaxadiol production from xylose by engineered Yarrowia lipolytica. Microb Cell Fact, 2019, 18: 83. DOI:10.1186/s12934-019-1136-7
|
| [70] |
Li DS, Wu YF, Zhang CB, et al. Production of triterpene ginsenoside compound K in the non-conventional yeast Yarrowia lipolytica. J Agric Food Chem, 2019, 67(9): 2581-2588. DOI:10.1021/acs.jafc.9b00009
|
| [71] |
Venter T, Kock JLF, Botes PJ, et al. Acetate enhances citric acid production by Yarrowia lipolytica when grown on sunflower oil. Syst Appl Microbiol, 2004, 27(2): 135-138. DOI:10.1078/072320204322881736
|
| [72] |
Liu XY, Lv JS, Xu JX, et al. Citric acid production in Yarrowia lipolytica SWJ-1b yeast when grown on waste cooking oil. Appl Biochem Biotechnol, 2015, 175(5): 2347-2356. DOI:10.1007/s12010-014-1430-0
|
| [73] |
Papanikolaou S, Galiotou-Panayotou M, Fakas S, et al. Citric acid production by Yarrowia lipolytica cultivated on olive-mill wastewater-based media. Bioresour Technol, 2008, 99(7): 2419-2428. DOI:10.1016/j.biortech.2007.05.005
|
| [74] |
Sarris D, Rapti A, Papafotis N, et al. Production of added-value chemical compounds through bioconversions of olive-mill wastewaters blended with crude glycerol by a Yarrowia lipolytica strain. Molecules, 2019, 24(2): 222. DOI:10.3390/molecules24020222
|
| [75] |
Ledesma-Amaro R, Lazar Z, Rakicka M, et al. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng, 2016, 38: 115-124. DOI:10.1016/j.ymben.2016.07.001
|
| [76] |
Zhang J, Zhang B, Wang DM, et al. Improving xylitol production at elevated temperature with engineered Kluyveromyces marxianus through over-expressing transporters. Bioresour Technol, 2015, 175: 642-645. DOI:10.1016/j.biortech.2014.10.150
|
| [77] |
Junior WGM, Pacheco TF, Trichez D, et al. Xylitol production on sugarcane biomass hydrolysate by newly identified Candida tropicalis JA2 strain. Yeast, 2019, 36(5): 349-361. DOI:10.1002/yea.3394
|
| [78] |
Jeon WY, Yoon BH, Ko BS, et al. Xylitol production is increased by expression of codon-optimized Neurospora crassa xylose reductase gene in Candida tropicalis. Bioprocess Biosyst Eng, 2012, 35(1/2): 191-198.
|
| [79] |
Veluturla S, Archna N, Rao DS, et al. Catalytic valorization of raw glycerol derived from biodiesel: a review. Biofuels, 2018, 9(3): 305-314. DOI:10.1080/17597269.2016.1266234
|
| [80] |
Mirończuk AM, Rzechonek DA, Biegalska A, et al. A novel strain of Yarrowia lipolytica as a platform for value-added product synthesis from glycerol. Biotechnol Biofuels, 2016, 9: 180. DOI:10.1186/s13068-016-0593-z
|
| [81] |
Rzechonek DA, Dobrowolski A, Rymowicz W, et al. Aseptic production of citric and isocitric acid from crude glycerol by genetically modified Yarrowia lipolytica. Bioresour Technol, 2019, 271: 340-344. DOI:10.1016/j.biortech.2018.09.118
|
| [82] |
Carly F, Vandermies M, Telek S, et al. Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metab Eng, 2017, 42: 19-24. DOI:10.1016/j.ymben.2017.05.002
|
| [83] |
Kata I, Semkiv MV, Ruchala J, et al. Overexpression of the genes PDC1 and ADH1 activates glycerol conversion to ethanol in the thermotolerant yeast Ogataea (Hansenula) polymorpha. Yeast, 2016, 33(8): 471-478. DOI:10.1002/yea.3175
|
| [84] |
Lazar Z, Walczak E, Robak M. Simultaneous production of citric acid and invertase by Yarrowia lipolytica SUC+ transformants. Bioresour Technol, 2011, 102(13): 6982-6989. DOI:10.1016/j.biortech.2011.04.032
|
| [85] |
Rakicka M, Lazar Z, Dulermo T, et al. Lipid production by the oleaginous yeast Yarrowia lipolytica using industrial by-products under different culture conditions. Biotechnol Biofuels, 2015, 8: 104. DOI:10.1186/s13068-015-0286-z
|
| [86] |
Galindo-Leva LÁ, Hughes SR, López-Núñez JC, et al. Growth, ethanol production, and inulinase activity on various inulin substrates by mutant Kluyveromyces marxianus strains NRRL Y-50798 and NRRL Y-50799. J Ind Microbiol Biotechnol, 2016, 43(7): 927-939. DOI:10.1007/s10295-016-1771-5
|
| [87] |
Liu XY, Chi Z, Liu GL, et al. Inulin hydrolysis and citric acid production from inulin using the surface-engineered Yarrowia lipolytica displaying inulinase. Metab Eng, 2010, 12(5): 469-476. DOI:10.1016/j.ymben.2010.04.004
|
| [88] |
Zhang HY, Zhang LN, Chen HQ, et al. Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP: citrate lyase from Mus musculus. J Biotechnol, 2014, 192: 78-84. DOI:10.1016/j.jbiotec.2014.10.004
|
| [89] |
Xu P, Qiao KJ, Ahn WS, et al. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc Natl Acad Sci USA, 2016, 113(39): 10848-10853. DOI:10.1073/pnas.1607295113
|
| [90] |
Zhou JW, Yin XX, Madzak C, et al. Enhanced α- ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J Biotechnol, 2012, 161(3): 257-264. DOI:10.1016/j.jbiotec.2012.05.025
|
| [91] |
Muhammad A, Feng XD, Rasool A, et al. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol Adv, 2020, 43: 107555. DOI:10.1016/j.biotechadv.2020.107555
|
| [92] |
Yang X, Nambou K, Wei LJ, et al. Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour Technol, 2016, 216: 1040-1048. DOI:10.1016/j.biortech.2016.06.028
|
| [93] |
Ignea C, Pontini M, Maffei ME, et al. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth Biol, 2014, 3(5): 298-306. DOI:10.1021/sb400115e
|
| [94] |
Matthäus F, Ketelhot M, Gatter M, et al. Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl Environ Microbiol, 2014, 80(5): 1660-1669. DOI:10.1128/AEM.03167-13
|
| [95] |
Liu H, Marsafari M, Wang F, et al. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab Eng, 2019, 56: 60-68. DOI:10.1016/j.ymben.2019.08.017
|
| [96] |
Liu H, Wang F, Deng L, et al. Genetic and bioprocess engineering to improve squalene production in Yarrowia lipolytica. Bioresour Technol, 2020, 317: 123991. DOI:10.1016/j.biortech.2020.123991
|
| [97] |
Dmytruk KV, Yatsyshyn VY, Sybirna NO, et al. Metabolic engineering and classic selection of the yeast Candida famata (Candida flareri) for construction of strains with enhanced riboflavin production. Metab Eng, 2011, 13(1): 82-88. DOI:10.1016/j.ymben.2010.10.005
|
| [98] |
Seip J, Jackson R, He HX, et al. Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica. Appl Environ Microbiol, 2013, 79(23): 7360-7370. DOI:10.1128/AEM.02079-13
|
| [99] |
Wang ZP, Xu HM, Wang GY, et al. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim Biophys Acta, 2013, 1831(4): 675-682. DOI:10.1016/j.bbalip.2012.12.010
|
| [100] |
Sharma NK, Behera S, Arora R, et al. Enhancement in xylose utilization using Kluyveromyces marxianus NIRE-K1 through evolutionary adaptation approach. Bioprocess Biosyst Eng, 2016, 39(5): 835-843. DOI:10.1007/s00449-016-1563-3
|
| [101] |
Cheng HR, Lv JY, Wang HW, et al. Genetically engineered Pichia pastoris yeast for conversion of glucose to xylitol by a single-fermentation process. Appl Microbiol Biotechnol, 2014, 98(8): 3539-3552. DOI:10.1007/s00253-013-5501-x
|
| [102] |
Gassler T, Sauer M, Gasser B, et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat Biotechnol, 2020, 38(2): 210-216. DOI:10.1038/s41587-019-0363-0
|
| [103] |
Daskalaki A, Perdikouli N, Aggeli D, et al. Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl Microbiol Biotechnol, 2019, 103(20): 8585-8596. DOI:10.1007/s00253-019-10088-7
|
 2021, Vol. 37
2021, Vol. 37




