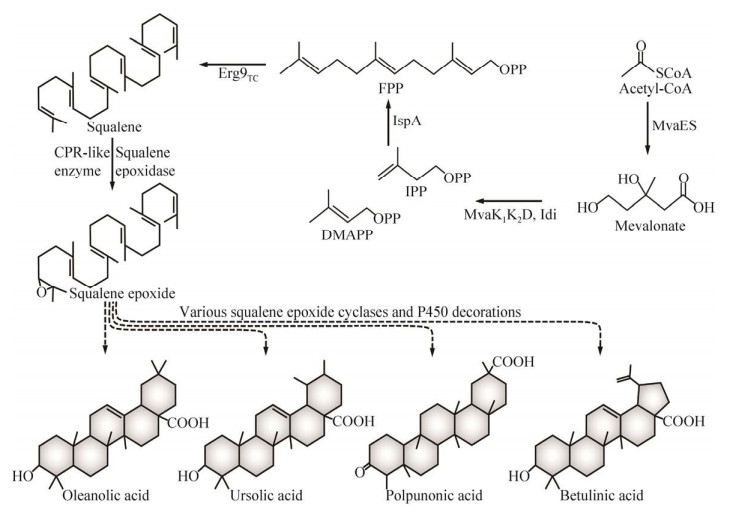
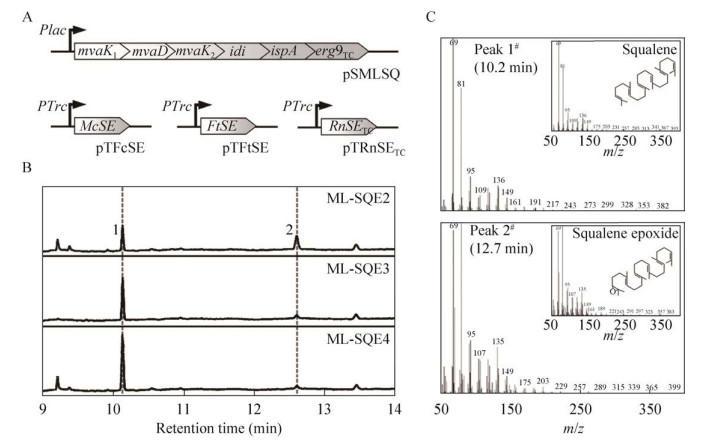
| [1] |
Liu H, Zhang Y, Sun SQ, et al. Efficacy of terpenoid in attenuating aortic atherosclerosis in apolipoprotein-E deficient mice: a meta-analysis of animal studies. Biomed Res Int, 2019, 2019: 2931831.
|
|
| [2] |
Shi Z, Chen Y, Lu C, et al. Resolving neuroinflammation, the therapeutic potential of the anti-malaria drug family of artemisinin. Pharmacol Res, 2018, 136: 172-180. DOI:10.1016/j.phrs.2018.09.002
|
|
| [3] |
Hua SY, Ma MY, Fei XY, et al. Glycyrrhizin attenuates hepatic ischemia-reperfusion injury by suppressing HMGB1-dependent GSDMD-mediated kupffer cells pyroptosis. Int Immunopharmacol, 2019, 68: 145-155. DOI:10.1016/j.intimp.2019.01.002
|
|
| [4] |
Zhang YY, Li LY, Qi C, et al. Glycyrrhizin alleviates Con A-induced hepatitis by differentially regulating the production of IL-17 and IL-25. Biomed Pharmacother, 2019, 110: 692-699. DOI:10.1016/j.biopha.2018.12.025
|
|
| [5] |
Kashyap D, Sharma A, Tuli HS, et al. Molecular targets of celastrol in cancer: recent trends and advancements. Crit Rev Oncol, 2018, 128: 70-81. DOI:10.1016/j.critrevonc.2018.05.019
|
|
| [6] |
Venkatesha SH, Dudics S, Astry B, et al. Control of autoimmune inflammation by celastrol, a natural triterpenoid. Pathog Dis, 2016, 74(6): ftw059. DOI:10.1093/femspd/ftw059
|
|
| [7] |
Cao L, Zhang X, Cao FF, et al. Inhibiting inducible miR-223 further reduces viable cells in human cancer cell lines MCF-7 and PC3 treated by celastrol. BMC Cancer, 2015, 15: 873. DOI:10.1186/s12885-015-1909-2
|
|
| [8] |
张倩茹, 高颖, 李群芳, 等. 杜仲叶环烯醚萜类化合物提取工艺研究. 安徽农业科学, 2010, 38(8): 4051-4052. Zhang QR, Gao Y, Li QF, et al. Study on the extraction of iridoids from Eucommia ulmoids leaves. J Anhui Agric Sci, 2010, 38(8): 4051-4052 (in Chinese). DOI:10.3969/j.issn.0517-6611.2010.08.081
|
|
| [9] |
常春, 赵亚南, 曾爱国. 环糊精水提法提取积雪草中的三萜类化合物. 西北药学杂志, 2018, 33(4): 432-435. Chang C, Zhao YN, Zeng AG. Extraction of triterpenoids from Centella asiatica by cyclodextrin aqueous extraction. Northwest Pharm J, 2018, 33(4): 432-435 (in Chinese). DOI:10.3969/j.issn.1004-2407.2018.04.002
|
|
| [10] | |
|
| [11] |
Surendra K, Corey EJ. A short enantioselective total synthesis of the fundamental pentacyclic triterpene lupeol. J Am Chem Soc, 2009, 131(39): 13928-13929. DOI:10.1021/ja906335u
|
|
| [12] |
Yang JM, Zhao G, Sun YZ, et al. Bio-isoprene production using exogenous MVA pathway and isoprene synthase in Escherichia coli. Bioresour Technol, 2012, 104: 642-647. DOI:10.1016/j.biortech.2011.10.042
|
|
| [13] |
Wang CL, Zada B, Wei GY, et al. Metabolic engineering and synthetic biology approaches driving isoprenoid production in Escherichia coli. Bioresour Technol, 2017, 241: 430-438. DOI:10.1016/j.biortech.2017.05.168
|
|
| [14] |
Wang CL, Yoon SH, Jang HJ, et al. Metabolic engineering of Escherichia coli for α-farnesene production. Metab Eng, 2011, 13(6): 648-655. DOI:10.1016/j.ymben.2011.08.001
|
|
| [15] |
Anthony JR, Anthony LC, Nowroozi F, et al. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4, 11-diene. Metab Eng, 2009, 11(1): 13-19. DOI:10.1016/j.ymben.2008.07.007
|
|
| [16] |
Huang QL, Roessner CA, Croteau R, et al. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg Med Chem, 2001, 9(9): 2237-2242. DOI:10.1016/S0968-0896(01)00072-4
|
|
| [17] |
Yang JM, Guo LZ. Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb Cell Fact, 2014, 13(1): 160. DOI:10.1186/s12934-014-0160-x
|
|
| [18] |
金应福, 韩莉, 张莎莎, 等. 通过番茄红素环化酶的优化构建β-胡萝卜素高产菌株. 生物工程学报, 2017, 33(11): 1814-1826. Jin YF, Han L, Zhang SS, et al. Construction of high-yield strain by optimizing lycopene cyclase for β-carotene production. Chin J Biotech, 2017, 33(11): 1814-1826 (in Chinese).
|
|
| [19] |
Yoon SH, Lee SH, Das A, et al. Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J Biotechnol, 2009, 140(3-4): 218-226. DOI:10.1016/j.jbiotec.2009.01.008
|
|
| [20] |
Ghimire GP, Lee HC, Sohng JK. Improved squalene production via modulation of the methylerythritol 4-phosphate pathway and heterologous expression of genes from Streptomyces peucetius ATCC 27952 in Escherichia coli. Appl Environ Microbiol, 2009, 75(22): 7291-7293. DOI:10.1128/AEM.01402-09
|
|
| [21] |
Katabami A, Li L, Iwasaki M, et al. Production of squalene by squalene synthases and their truncated mutants in Escherichia coli. J Biosci Bioeng, 2015, 119(2): 165-171. DOI:10.1016/j.jbiosc.2014.07.013
|
|
| [22] |
Xu W, Yao J, Liu LJ, et al. Improving squalene production by enhancing the NADPH/NADP + ratio, modifying the isoprenoid-feeding module and blocking the menaquinone pathway in Escherichia coli. Biotechnol Biofuels, 2019, 12: 68. DOI:10.1186/s13068-019-1415-x
|
|
| [23] |
Meng YH, Shao XX, Wang Y, et al. Extension of cell membrane boosting squalene production in the engineered Escherichia coli. Biotechnol Bioeng, 2020, 117(11): 3499-3507. DOI:10.1002/bit.27511
|
|
| [24] |
Yang LY, Wang CL, Zhou J, et al. Combinatorial engineering of hybrid mevalonate pathways in Escherichia coli for protoilludene production. Microb Cell Fact, 2016, 15: 14. DOI:10.1186/s12934-016-0409-7
|
|
| [25] | |
|
| [26] | |
|
| [27] | |
|
| [28] |
Campbell ZT, Baldwin TO. Fre is the major flavin reductase supporting bioluminescence from Vibrio harveyi luciferase in Escherichia coli. J Biol Chem, 2009, 284(13): 8322-8328. DOI:10.1074/jbc.M808977200
|
|
| [29] | |
|
| [30] |
Driggers CM, Dayal PV, Ellis HR, et al. Crystal structure of Escherichia coli SsuE: defining a general catalytic cycle for FMN reductases of the flavodoxin-like superfamily. Biochemistry, 2014, 53(21): 3509-3519. DOI:10.1021/bi500314f
|
|
| [31] |
Andrade SL, Patridge EV, Ferry JG, et al. Crystal structure of the NADH: quinone oxidoreductase WrbA from Escherichia coli. J Bacteriol, 2007, 189(24): 9101-9107. DOI:10.1128/JB.01336-07
|
|
| [32] |
Sakakibara J, Watanabe R, Kanai Y, et al. Molecular cloning and expression of rat squalene epoxidase. J Biol Chem, 1995, 270(1): 17-20. DOI:10.1074/jbc.270.1.17
|
|
| [33] |
Nakano C, Motegi A, Sato T, et al. Sterol biosynthesis by a prokaryote: first in vitro identification of the genes encoding squalene epoxidase and lanosterol synthase from Methylococcus capsulatus. Biosci Biotechnol Biochem, 2007, 71(10): 2543-2550. DOI:10.1271/bbb.70331
|
|
| [34] |
Nagumo A, Kamei T, Sakakibara J, et al. Purification and characterization of recombinant squalene epoxidase. J Lipid Res, 1995, 36(7): 1489-1497. DOI:10.1016/S0022-2275(20)39736-4
|
|
| [35] |
Dinkova-Kostova AT, Talalay P. NAD(P)H: quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys, 2010, 501(1): 116-123. DOI:10.1016/j.abb.2010.03.019
|
|
| [36] |
Friedrich T, Dekovic DK, Burschel S. Assembly of the Escherichia coli NADH: ubiquinone oxidoreductase (respiratory complexⅠ). Biochim Biophys Acta, 2016, 1857(3): 214-223. DOI:10.1016/j.bbabio.2015.12.004
|
|
| [37] |
Li DS, Zhang Q, Zhou ZJ, et al. Heterologous biosynthesis of triterpenoid dammarenediol-Ⅱ in engineered Escherichia coli. Biotechnol Lett, 2016, 38(4): 603-609. DOI:10.1007/s10529-015-2032-9
|
|
| [38] |
Arnesen JA, Kildegaard KR, Cernuda Pastor M, et al. Yarrowia lipolytica strains engineered for the production of terpenoids. Front Bioeng Biotechnol, 2020, 8: 945. DOI:10.3389/fbioe.2020.00945
|
|
 2021, Vol. 37
2021, Vol. 37