中国科学院微生物研究所、中国微生物学会主办
文章信息
- 熊婉, 段燕文, 颜晓晖, 黄勇
- Xiong Wan, Duan Yanwen, Yan Xiaohui, Huang Yong
- 途径特异性调控因子介导的链霉菌来源天然产物的产量提升
- Improvement of natural product production in Streptomyces by manipulating pathway-specific regulators
- 生物工程学报, 2021, 37(6): 2127-2146
- Chinese Journal of Biotechnology, 2021, 37(6): 2127-2146
- 10.13345/j.cjb.200777
-
文章历史
- Received: December 2, 2020
- Accepted: March 5, 2021
- Published: March 18, 2021
2. 组合生物合成与天然产物药物湖南省工程研究中心,湖南 长沙 410205;
3. 新药组合生物合成国家地方联合工程研究中心,湖南 长沙 410205;
4. 天津中医药大学省部共建组分中药国家重点实验室,天津 301617
2. Hunan Engineering Research Center of Combinatorial Biosynthesis and Natural Product Drug Discovery, Changsha 410205, Hunan, China;
3. National Engineering Research Center of Combinatorial Biosynthesis for Drug Discovery, Changsha 410205, Hunan, China;
4. State Key Laboratory of Component-based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, China
链霉菌产生的天然产物是抗生素的主要来源,如链霉素、四环素和万古霉素。目前临床上超过一半抗生素来源于链霉菌[1]。链霉菌也是重要的抗肿瘤药物(放线菌素、多柔比星、博来霉素)、免疫抑制剂(雷帕霉素) 和杀虫剂(阿维菌素) 的产生菌[2-3]。21世纪初报道的天蓝色链霉菌Streptomyces coelicolor和阿维链霉菌Streptomyces avermitilis的全基因组序列揭示了链霉菌作为“抗生素工厂”的遗传物质基础,即它们分别含有22个和38个天然产物生物合成基因簇(Biosynthetic gene clusters,BGCs)[4-5]。近20年来,对链霉菌的基因组分析发现每个菌株平均有20–40个天然产物生物合成基因簇。这些研究揭示了包括链霉菌在内的微生物具有巨大的生物合成潜力,并开启了微生物天然产物发现的“后基因组时代”。人们基于对各类天然产物生物合成机制及其基因簇的认知,从基因组出发可以有针对性地发现目标菌株中结构新颖的天然产物。与利用传统的基于发酵和活性筛选的微生物天然产物发现方法相比,基因组挖掘技术可提高在微生物中发现新颖化合物的成功率[6-7]。
链霉菌属于高GC含量的革兰氏阳性丝状放线菌,其特定天然产物的产生和产量高低与菌株本身复杂的形态分化机制和代谢网络密切相关。链霉菌来源的天然产物由链霉菌在特定的生长条件下,受到自身形态分化或环境胁迫的影响而产生,可以用于化学防御或作为信号分子用于物种间的交流。因此,天然产物的产生及产量通常受到多层次、多维度的精准调控,例如全局性调控因子(Pleiotropic regulators) 和多种途径特异性调控因子(Pathway-specific regulators),以及生物合成的前体供应和生物合成基因簇的拷贝数等[8-9]。
合成生物学的兴起为链霉菌来源的天然产物产量提升提供了一个强有力的策略。例如,Smanski等敲除普拉特链霉菌Streptomyces platensis MA7327中的调控基因ptmR1,构建了二萜类抗生素平板霉素(Platensimycin) 和平板素(Platencin) 的高产菌株,其产量分别达到323 mg/L和255 mg/L,约为野生菌株的100倍[10];在普拉特链霉菌MA7339中,敲除同源基因ptnR1将平板素的产量从0.05 mg/L提升至22 mg/L[11]。增加生物合成基因簇的拷贝数可以提升链霉菌来源天然产物的产量。例如,Murakami等通过DNA重组的方法在天蓝色链霉菌中将放线紫红素的BGC扩增了4–12倍,从而使得放线紫红素产量提升了20倍以上[12]。敲除天然产物竞争性代谢途径以及增加前体供应都是提升链霉菌来源天然产物的有效方法。盐霉素是由白色链霉菌Streptomyces albus产生的聚酮类化合物,Lu等通过敲除其竞争性聚酮BGC后得到了突变株,其盐霉素产量达5.7 g/L;在此菌株中过表达巴豆酰辅酶A增加了盐霉素生物合成过程中的前体供应,最终将盐霉素的产量提升至6.6 g/L[13]。Wang等通过过表达脂酰辅酶A合成酶(Fatty acyl-CoA synthetase) 促进了胞内三酰甘油的分解代谢,提升了乙酰辅酶A的浓度,该策略大幅度提升了放线紫红素、杰多霉素B、土霉素或阿维菌素B1a的产量;进一步在180 L的小试发酵中将工业菌种阿维链霉菌中阿维菌素B1a的产量从6.2 g/L提升到9.31 g/L[14-16]。上述研究说明了对链霉菌代谢网络及调控机制的研究将加速其产生的天然产物产量的提升。
因此,国内外学者近期分别综述了链霉菌中的转录因子[17]、新颖的合成生物学策略对链霉菌等放线菌的产量提升[8]、放线菌中抗生素生物合成的复杂级联调控[18]以及与链霉菌中天然产物生物合成相关的信号分子、调控基因及反馈调控机制等的影响[19-20]。但是通过在中国知网上用“链霉菌”和“调控”等关键词搜索,近年来综述链霉菌调控的中文文章很少。本文在Web of Science数据库分别以“Streptomyces”和“Regulatory gene”、“Regulators”、“Regulation”为关键词检索了相关研究论文,并将主要综述2015–2020年来通过对链霉菌中途径特异性调控因子的操控来提升其活性天然产物产量的研究进展。
1 链霉菌中的调控基因基于调控基因的遗传操作是菌株改良的有效工具,对调控基因的深入了解是提升抗生素产量或者激活沉默基因簇的先决条件。在微生物天然产物生物合成中调控基因通常分为两类:一类为途径特异性调控基因,它通常位于特定的BGCs内,影响着簇内基因的表达;另一类为全局性调控基因,通常位于生物合成基因簇的外部,它控制多种代谢途径比如链霉菌的初级代谢以及形态分化,可能不直接影响特定BGCs的表达[21]。在天然产物生物合成的过程中,微生物通过全局性调控因子对环境中的营养及环境压力信号作出响应,例如磷酸盐浓度、碳氮源、几丁质或N-乙酰氨基葡萄糖,以及微生物细胞壁损伤、热休克或pH胁迫等,调控下游基因的表达进而影响途径特异性调控基因,并控制初级代谢水平[22-23]。在特定的培养条件下,信号分子、调控因子和反馈调节之间相互作用决定了次级代谢产物的产生及其产量的高低[20](图 1)。链霉菌来源天然产物的产生受途径特异性调控基因和全局性调控基因的控制,这也为它们产量的提升提供了两种不同的策略,即操纵全局调控基因以增加多种次级代谢产物产量,以及操纵途径特异性调控基因提升特定天然产物产量[24-25]。途径特异性调控基因与天然产物生物合成基因的表达水平关系密切,这使其成为提高特定天然产物产量的研究目标[26-27]。
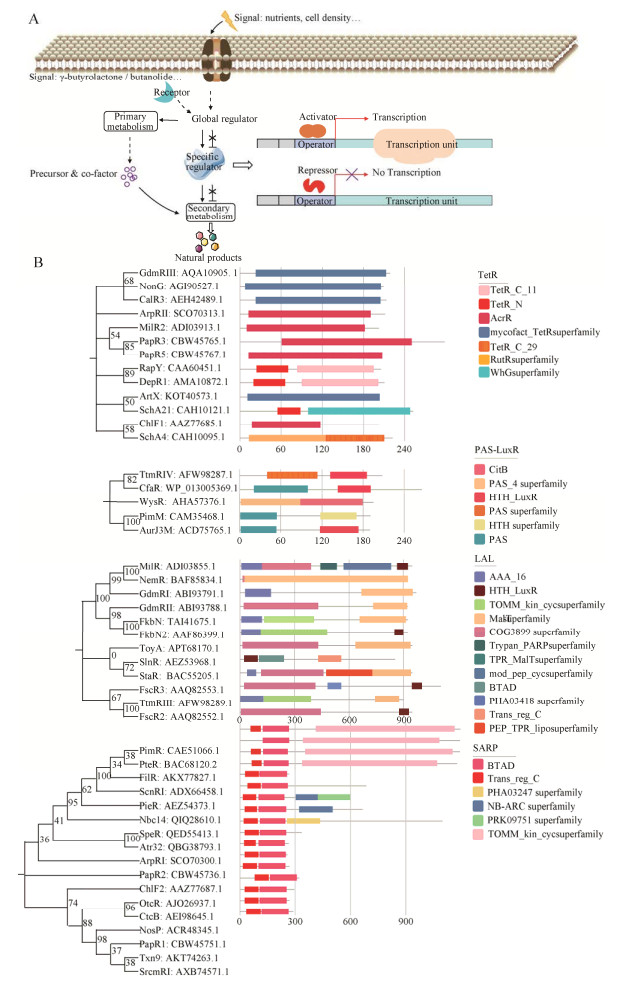
|
| 图 1 链霉菌来源天然产物的调控 Fig. 1 Regulation of the biosynthesis of Streptomyces natural products. (A) The regulatory cascades of natural product production in Streptomyces. (B) Phylogenetic tree and domain analysis of pathway-specific regulators in Streptomyces. |
| |
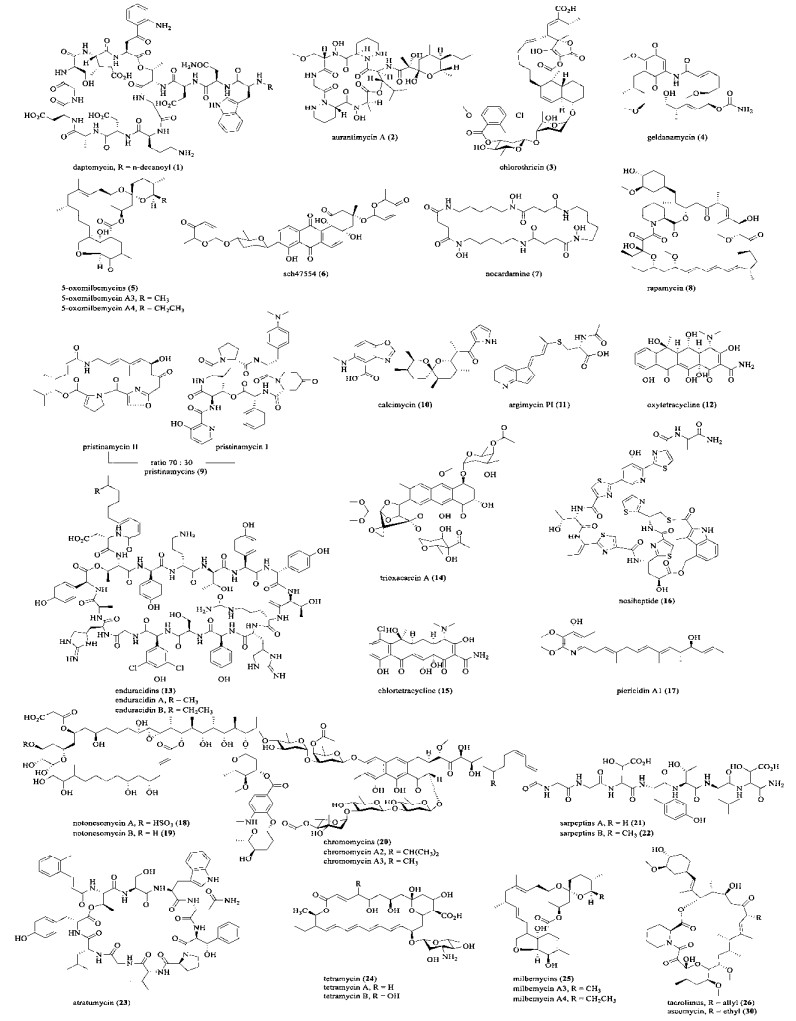
|
| 图 2 由TetR、SARP家族调控因子调控产生的天然产物化学结构 Fig. 2 Structures of natural products regulated by TetR and SARP family regulators. |
| |
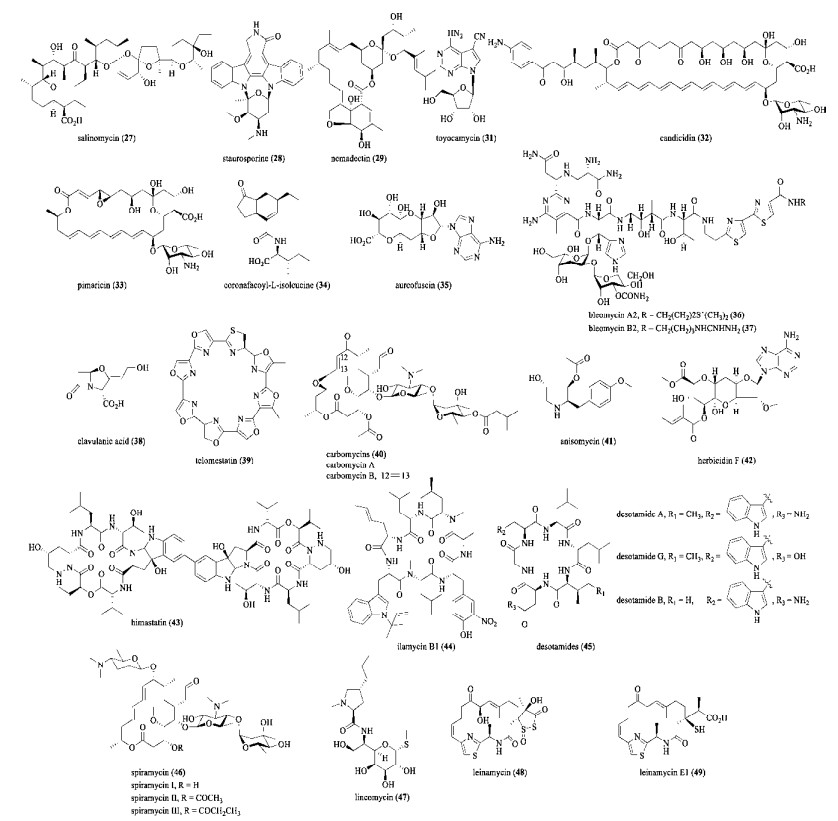
|
| 图 3 由LuxR和其他家族调控因子调控产生的天然产物化学结构 Fig. 3 Structures of natural products regulated by LuxR and other family regulators. |
| |
链霉菌调控网络平衡其初级和次级代谢,而途径特异性调控因子则是其复杂调控网络中的关键节点[25]。大部分参与转录调控的途径特异性调控因子包含两个蛋白结构域,分别为信号接收结构域(Signal receiving domain) 和DNA结合结构域(DNA-binding domain,DBD)[28]。根据DNA结合结构域之间的序列相似性、氨基酸序列的排列以及结构和功能等标准对链霉菌中途径特异性调控因子进行分类,通常将它们分为TetR、LuxR、SARP (Streptomyces antibiotic regulatory protein)、LysR、MarR等家族[18]。TetR家族转录调控因子(TetR family of regulators,TRFs) 由一个N端DBD (约50个氨基酸) 和一个较长的C端结构域组成。例如,RapY和DepR1调控因子由大小约50个氨基酸的TetR_N结构域和约100个氨基酸的TetR_C_11结构域组成(图 1B和《生物工程学报》网络版附件支持材料)。C端结构域通常与一个或多个配体相互作用,从而改变调控因子与DNA的结合能力。部分TetR家族转录调控因子属于负调控蛋白,它们通过DBD介导,与操纵子结合后抑制目标基因转录[29-31]。TetR调控因子也参与调控和抗生素的外排、氨基酸代谢以及细胞渗透压相关基因的表达等[32]。
大部分LuxR家族转录调控因子与微生物群体感应相关,在革兰氏阴性菌中居多,但其在链霉菌中可以调控天然产物的生物合成[33]。LuxR通常少于250个氨基酸,由一个信号接收结构域和一个DNA结合结构域组成。与TetR调控因子不同,LuxR通常N端是与群体感应相关的自诱导分子(Autoinducer,AI) 结合区域或响应调控区域,而C端利用典型的螺旋-转角-螺旋(Helix-turn-helix,HTH) 结构域与DNA结合[17]。部分LuxR调控因子还包括其他与信号识别和转导相关的结构域,例如PAS-LuxR亚家族,包括TtmRIV、CfaR、AuJ3M、MilR、FscR3等调控因子[34-35]。LAL调控因子(Large ATP-binding regulators of the LuxR family) 是LuxR调控因子的另一个亚家族,其原型来自于大肠杆菌Escherichia coli中参与麦芽糊精吸收和分解代谢的MalT。编码LAL型调控因子的基因广泛分布于链霉菌,尤其是在I型聚酮类化合物的BGC中[25]。该亚家族与PAS-LuxR相比相对较大,约由900个氨基酸构成。在C末端是典型的LuxR家族的HTH结构域与DNA结合,而N端则包含一个ATP/GTP结合结构域,负责与诱导物、鼠李糖和ATP相互作用[36]。少数LAL调控因子的N端与DNA结合,如SlnR。SARP家族转录调控因子迄今只在放线菌中发现,而其中大多数存在于链霉菌中[17]。SARP蛋白通常包括两个代表性结构域,即与DNA结合的N端侧翼HTH结构域(Winged-HTH,wHTH) 和C端细菌转录激活因子结构域(Bacterial transcription activation domain,BTAD)[25]。SARP-LAL类型调控因子比较独特,除了典型的wHTH结构域外,其C端为一个LAL类型结构域,包括PimR、ScnRI、PteR、FilR等。
LysR家族转录调控因子大小约为310–325个氨基酸,在N末端具有与DNA结合的保守HTH结构域,C末端则是与底物或者诱导物结合的调节域[21],包括Tcs7、IlaA、ClaR等调控因子。MarR家族转录调控因子在链霉菌中含量丰富,平均每个基因组中含有50个MarR型调控因子。尽管数量众多,对这类调控蛋白的研究却相对甚少[17]。MarR家族调控因子中央区域具有保守的wHTH结构域与DNA结合,包括Slinc377、CtcS等。由于前3种家族调控因子在途径特异性调控中较为常见,下文将对其进行重点介绍。
2 通过途径特异性调控因子的表达调控提升天然产物产量 2.1 TetR家族调控因子TetR家族调控因子数量庞大,目前在链霉菌中已发现超过100个TetR家族调控因子[37]。在玫瑰孢链霉菌Streptomyces roseosporus SW0702中,两个TetR家族调控因子AtrA、DepR1作为正向调控因子促进达托霉素(Daptomycin,1) 生物合成。其中AtrA为全局性调控因子,而DepR1为途径特异性调控因子。在钙离子的存在下,1通过与革兰氏阳性耐药菌的细胞膜结合发挥抗菌作用,1对耐万古霉素的金黄色葡萄球菌Staphylococcus aureus、耐甲氧西林的金黄色葡萄球菌、耐青霉素的肺炎链球菌Streptococcus pneumoniae和耐万古霉素的肠球菌Enterococcus均有很强的抗菌活性[38]。自2003年上市以来,达托霉素的市场应用前景广阔,然而其产量不高,需要进一步提升其产量以降低其生产成本[39]。Yuan等在玫瑰孢链霉菌中过表达depR1将达托霉素的产量提升了41%,达到474 mg/L (表 1)[40]。Aurantimycin (2) 是由橘橙链霉菌Streptomyces aurantiacus产生的脂肽类抗肿瘤抗生素,其生物合成途径和调控机制的研究较少。Zhao等过表达artX将2的产量提升到了原始菌株的2倍,达到200 mg/L[41]。在链霉菌中分别过表达chlF1或gdmRIII,提升了chlorothricin (3) 或geldanamycin (4) 的产量[42-43]。米尔贝肟(Milbemycin oxime) 是米尔贝霉素A3/A4 (Milbemycin A3/A4) 的肟衍生物,对预防和控制常见寄生虫疾病有良好的效果。化合物5-酮米尔贝霉素(Oxomilbemycin A3/A4,5) 是米尔贝肟的合成中间体。Wei等过表达吸水链霉菌Streptomyces hygroscopicus中的milR2,不仅缩短了5在摇瓶和发酵罐中的发酵周期,并将5的产量较出发菌株提升34.4%[44],达到约3.6 g/L。米尔贝肟的化学合成途径以CrO3为氧化剂,将米尔霉素A3/A4氧化成5。该反应效率低下,而且CrO3的使用不利于环境保护,因此利用微生物发酵高产5不仅具有一定的经济价值,对环境保护也有重要意义。Fidan等发现调控基因schA4和schA21对链霉菌Streptomyces sp. SCC-2136中Sch47554 (6) 的生物合成具有不同的调控作用:敲除schA4可以将6的产量较野生菌株提升4.2倍;敲除schA21后6的产量大大降低,而过表达schA21将6的产量较野生菌株也提升了4倍[45]。Park等通过在白色链霉菌中异源表达nonactin的生物合成基因簇,发现其中的nonG调控基因可以激活原来沉默的铁载体基因簇,产生了nocardamine (7)[46]。
| Family (Modea) | Regulator | Strains | Antibiotics | Yield (Fold/(mg/L)) | References |
| TetR (+) | DepR1 | S. roseosporus | Daptomycin (1) | ~1.4/474 | [40] |
| ArtX | S. aurantiacus | Aurantimycin A (2) | 2/~200 | [41] | |
| ChlF1 | S. autolyticus | Chlorothricin (3) | – | [42] | |
| GdmRIII | S. autolyticus | Geldanamycin (4) | – | [43] | |
| MilR2 | S. hygroscopicus | 5-oxomilbemycin A3/A4 (5) | ~1.3/3 600 | [44] | |
| SchA21 | Streptomyces sp. SCC-2136 | Sch47554 (6) | ~3.9/26.2 | [45] | |
| NonG | S. albus | Nocardamine (7) | activated | [46] | |
| TetR (-) | RapY | S. rapamycinicus | Rapamycin (8) | 3.7/26.7 | [47] |
| PapR3 | S. pristinaespiralis | Pristinamycins (9) | ~1.5/– | [48] | |
| PapR5 | ~3/– | ||||
| CalR3 | S. chartreusis | Calcimycin (10) | ~8/– | [49] | |
| ArpRII | S. argillaceus | Argimycin PI (11) | 1.6~3.6/– | [50] | |
| SchA4 | Streptomyces sp. SCC-2136 | Sch47554 (6) | ~4/27.9 | [45] | |
| SARP (+) | PapR1 PapR2 | S. pristinaespiralis | Pristinamycins (9) | ~2/– ~2/– | [51] |
| OtcR | S. rimosus | Oxytetracycline (12) | ~6.5/6 240 | [52] | |
| Orf22 | S. fungicidicus | Enduracidins (13) | ~4.0/2.9 | [53] | |
| Txn9 | S. bottropensis | Trioxacarcin A (14) | – | [54] | |
| CtcB | S. aureofaciens | Chlortetracycline (15) | – | [55] | |
| NosP | S. actuosus | Nosiheptide (16) | – | [56] | |
| ArpRI | S. argillaceus | Argimycin PI (11) | 1.2~1.9/– | [50] | |
| PieR | S. piomogeues | Piericidin A1 (17) | 2.3/– | [57] | |
| ChlF2 | S. antibioticus DSM 40725 | Chlorothricin (3) | 7.6/– | [58] | |
| Nbc14 | Streptomyces sp. A793 | Notonesomycin A (18) Notonesomycin B (19) | 18/– 3/– | [59] | |
| SrcmRI | S. reseiscleroticus | Chromomycins (20) | activatedb/22.1 | [60] | |
| SpeR | Streptomyces sp. KO-7888 | Sarpeptin A (21) Sarpeptin B (22) | activated | [61] | |
| SARP (-) | Atr32 | S. atratus SCSIO ZH16NS-80S | Atratumycin (23) | 2.3/823 | [62] |
| LAL (+) | TtmRIII | S. ahygroscopicus | Tetramycin (24) | ~0.8(decreasedc)/– | [63] |
| MilR | S. bingchenggensis | Milbemycin A3/A4 (25) | ~1.4/4 069 | [64] | |
| FkbN | S. tsukubaensis | Tacrolimus (26) | ~1.8/252.2 | [65] | |
| SlnR | S. albus | Salinomycin (27) | ~1.3/– | [66] | |
| StaR | S. fradiae | Staurosporine (28) | 5.2/– | [67] | |
| GdmRI | S. hygroscopicus | Geldanamycin (4) | ~1.3/– | [68] | |
| GdmRII | ~1.3/– | ||||
| NemR | S. cyaneogriseus | Nemadectin (29) | 1.8/595 | [69] | |
| FkbN2 | S. hygroscopicus var. ascomyceticus | Ascomycin (30) | 4/1 800 | [70] | |
| ToyA | S. diastatochromogenes | Toyocamycin (31) | 2/456.3 | [71] | |
| FscR2, FscR3 | Streptomyces sp. FR-008. | Candicidin (32) | – | [72] | |
| PAS-LUXR (+) | PimM | S. natalensis | Pimaricin (33) | 1.2/– | [36] |
| TtmRIV | S. ahygroscopicus | Tetramycin A (24) | 3.3/1 334 | [63] | |
| CfaR | S. scabies | Coronafacoyl-L-isoleucine (34) | 10/- | [73] | |
| AurJ3M | S. aureofuscus | Aureofuscin (35) | – | [74] | |
| aThe following modes of regulation are indicated: +, activator; -, repressor. bActivation of silent BGCs. c decreased yields compared to the original strain. | |||||
上述TetR类型调控因子在链霉菌来源天然产物的生物合成中起着正调控的作用,部分TetR调控因子可以抑制天然产物的产生,例如RapY、PapR3和PapR3、CalR3等。Yoo等通过敲除链霉菌S. rapamycinicus中的调控基因rapY,将雷帕霉素(Rapamycin,8) 的产量提升至26.7 mg/L,为野生菌株的3.7倍[47]。在始旋链霉菌Streptomyces pristinaespiralis中,普那霉素(Pristinamycin,9) 的产生受到不同转录调控因子的级联调控,包括γ-丁内酯、反应调节因子以及6个途径特异性调控因子。Mast等通过敲除两个编码TetR类调控因子的基因papR3或papR5,将9的产量分别提升了1.5倍和3倍,而过表达两个编码SARP类调控因子的基因papR1和papR2将9的产量提升了1倍[48]。通过敲除calR3或arpRII调控基因,calcimycin (10) 或argimycins P (11) 产量均得到了提升,其中10在突变菌株中的产量约为原始菌株的8倍[49-50]。
值得注意的是,TetR类型调控因子不仅局限于控制特定次级代谢产物生物合成基因的表达,也通过调控初级代谢相关基因的表达来影响着次级代谢产物生物合成。例如,Lyu等最近发现阿维链霉菌中的AccR调控因子通过调控乙酰辅酶A羧化酶和支链氨基酸代谢的相关基因,来调节细胞内短链酰基辅酶A的水平,进而影响次级代谢产物的产生。敲除accR使阿维菌素B1a的产量比野生菌株中提高了1.6倍,而在阿维链霉菌A8中将阿维菌素B1a的产量提升了14.5%,达到7.76 g/L[51]。
2.2 SARP家族调控因子SARP类型调控因子通常通过直接激活链霉菌来源天然产物的生物合成基因或其他调控基因来发挥其功能[56]。在龟裂链霉菌Streptomyces rimosus中的土霉素(Oxytetracycline,12) 生物合成基因簇里,SARP家族的调控因子OtcR直接激活簇内5个启动子来正向调控12的生物合成。化合物12等四环素类抗生素能够抑制细菌蛋白质的合成,具有广谱的抗菌活性。土霉素的耐药性导致其临床应用减少,但在饲料禁抗以前,仍然被广泛用于动物饲料和水产养殖中,年生产超过5 000 t。Yin等在龟裂链霉菌中过表达otcR,将其产量提升了6.49倍,达到6.24 g/L[52]。Enduracidins (13) 是一类脂肽抗生素,对耐万古霉素肠球菌和耐甲氧西林金黄色葡萄球菌等革兰氏阳性菌具有良好的抑菌活性。Chen等通过在杀真菌素链霉菌Streptomyces fungicidicus中过表达簇内调控基因orf22,将enduracidin A和B的产量从0.74 mg/L提升至约2.93 mg/L[53]。过表达txn9、ctcB、nosP、arpRI、pieR或chlF2调控基因,分别提升了trioxacarcin A (14)、chlortetracycline (15)、nosiheptide (16)、argimycins P (11)、piericidin A1 (17) 和chlorothricin (3) 的产量。例如,3的产量较其野生菌株提高了7.6倍,而17的产量较野生菌株提高了2.3倍[50, 54-58]。Goh等利用CRISPR-Cas9介导的基因组编辑技术在链霉菌Streptomyces sp. A793中过表达调控基因nbc14,将notonesomycin A (18)和notonesomycin B (19)的产量分别提升了18倍和3倍[59]。
操纵途径特异性调控因子不仅能提升特定次级代谢产物产量,也是发现新天然产物的有效方法之一。Sun等发现在链霉菌S. reseiscleroticus中存在一个沉默的chromomycins (20) 生物合成基因簇;将该基因簇在异源宿主变青铅链霉菌Streptomyces livdans中进行异源表达并过表达调控基因srcmRI后,发现了chromomycin同系物的产生[60]。Koomsiri等在链霉菌Streptomyces sp. KO7888中发现一个沉默的编码非核糖体多肽类化合物的基因簇;过表达其调控基因speR后产生了两个新的脂肽sarpeptins A (21) 和sarpeptins B (22)[61]。少数链霉菌SARP类型调控因子抑制次级代谢产物的生物合成。例如,FarR4负调控淡紫灰链霉菌Streptomyces lavendulae中靛青的生物合成[75]。Yang等通过敲除暗黑链霉菌Streptomyces atratus中的调控基因atr32,将atratumycin (23) 的产量提升了2.3倍,达到823 mg/L[62]。另外,SARPs还可作为多效性调节蛋白,控制多种次级代谢产物的产生以及链霉菌孢子的形态分化[76]。例如,Santos-Beneit等发现AfsR通过与调控基因phoP的启动子结合,与PhoP和GlnR调控因子组成复杂的调控网络,共同参与调控天蓝色链霉菌中的初级与次级代谢,维持胞内氮和磷的代谢稳态[77];2018年,Ma等发现在链霉菌S. tsukubaensis中BulZ作为多效性调控因子,调节其孢子分化以及他克莫司的产生[78]。因此,SARP类型调控因子对链霉菌中天然产物的生物合成展现出复杂多样的调控模式。
2.3 LuxR家族调控因子在链霉菌中,超过20种LAL家族调控因子对天然产物的产生起着关键作用[67]。例如,tetramycin (24) 是由链霉菌S. ahygroscopicus产生的一类具有抗真菌作用的多烯大环内脂类抗生素,Cui等发现过表达ttmRI或ttmRIII后tetramycin的产量分别下降至野生菌株的75.7%和51.6%。作者推测产生这一异常现象的原因可能是这两个调控基因的表达量超过了阈值[63]。十六元环的大环内酯类化合物米尔贝霉素(Milbemycin, 25) 作为杀虫剂应用广泛。在冰城链霉菌Streptomyces bingchenggensis BC04中,25的产量约为2.95 g/L,通过过表达milR将其产量提升至4.07 g/L,降低了25的生产成本[64]。在工业菌株S. tsukubaensis L19中过表达fkbN和tcs7将tacrolimus (26) 的产量提升至272.1 mg/L,约为原始菌株的1.9倍[65]。通过过表达slnR或staR,分别将salinomycin (27) 或staurosporine (28) 的产量提升到野生菌株的1.25倍和5.2倍[66-67]。在吸水链霉菌中分别过表达gdmRI或gdmRII,将geldanamycin (4) 的产量提升了33%[68]。
Moxidectin是一种高效的杀虫剂,广泛用于治疗牛、羊和宠物常患的寄生虫疾病。Moxidectin的前体nemadectin (29) 是从蓝灰链霉菌Streptomyces cyaneogriseus发酵液中分离的16元环大环内酯。Li等利用hrdB启动子过表达nemR调控基因,将29的产量从331 mg/L提升至595 mg/L,较野生菌株提升了79.9 %[69]。长川霉素(Ascomycin,30) 是一种具有高免疫抑制活性的23元大环内酯类化合物。Zhang等在链霉菌S. tsukubaensis中过表达fkbN,将30的产量提升至1.80 g/L,较原始菌株提升4倍[70]。核苷类抗生素丰加霉素(Toyocamycin,31) 对多种植物病原真菌有较强的抗菌活性。在淀粉酶产色链霉菌Streptomyces diastatochromogenes中,Xu等利用不同强度的启动子SPL57、SPL21或PermE*分别过表达toyA,将31的产量分别提升至野生菌株的3倍、2倍和1.8倍[71]。
链霉菌Steptomyces sp. FR-008中多烯大环内酯杀念珠菌素(Candicidin,32) 的生物合成基因簇内包含4个相邻的调控基因,其中fscR1编码LuxR类型调控因子,而fscR2、fscR3和fscR4均编码LAL家族调控因子。这4个调控基因形成一个亚簇,具有较保守的调控机制,并分布在如两性霉素、制霉菌素等其他多个多烯大环内酯的生物合成基因簇中。Zhu等发现这些调控基因在32的生物合成过程中发挥着正向的调控作用,并组成了一个精细的调控网络(图 4):FscR2和FscR3位于调控网络的上游,FscR3正调节FscR2,而FscR2负调节FscR3,形成负反馈环路;在调控网络的下游,FscR1和FscR4可以相互正向调节;最后,FscR1也由FscR2和FscR3正向调节,FscR4由FscR2正向调控,而由FscR3负向调控。杀念珠菌素生物合成基因簇中的这个调控网络对于研究其他多烯大环内酯的生物合成的调控模型颇具借鉴意义[72]。
PAS-LuxR调控因子高度保守,目前已知的多烯大环内酯生物合成基因簇均包含一个编码PAS-LuxR类型调控因子的基因[79]。此类调控基因的原型是纳塔尔链霉菌Streptomyces natalensis中匹马霉素(Pimaricin, 33) 基因簇里的正调控基因pimM[80]。Santos-Aberturas等在链霉菌纳塔尔链霉菌中引入编码PAS-LuxR类型调控因子的外源基因,包括amphRIV、nysRIV或pteF到S. natalensis △pimM菌株中,33的产量得以恢复。这说明外源PAS-LuxR调节因子与PimM的功能类似,为提升多烯大环内酯的产量提供了一个有效的策略[81]。四霉素(Tetramycin,24) 是26元环四烯类抗生素,由四霉素A和B两部分组成,对多种植物病原菌有较高抑菌活性。Cui等通过在链霉菌S. ahygroscopicus中过表达ttmRIV,将四霉素A的产量较野生菌株提升了3.3倍,达到约1.33 g/L[65]。另外,分别过表达cfaR或aurJ3M调控基因,提升了coronafacoyl-L-isoleucine (34) 和aureofuscin (35) 的产量,而34产量在cfaR过表达菌株中是原始菌株的10倍[73-74]。
2.4 其他途径特异性调控因子除了较常见的TetR、SARP和LuxR类型调控因子外,其他类型的调控因子,如LysR、GntR和MarR等,也在链霉菌次级代谢产物的生物合成中起着重要作用,包括激活或者抑制特定天然产物的生物合成[19]。C-1027由烯二炔发色团和酸性辅基蛋白组成,具有优异的抗肿瘤活性,在我国已经进入Ⅱ期临床研究。在球孢链霉菌Streptomyces globisporus中,C-1027的产量较低((5.5±1.3) mg/L)[82]。C-1027的生物合成基因簇有4个调控基因,分别编码属于AraC/XylS家族的SgcR2、类StrR型调控因子SgcR1、类TylR型调控因子SgcR3以及新颖的调控因子SgcR。Chen等通过过表达sgcR1、sgcR2和sgcR3,以及敲除sgcR都提升了C-1027的产量。其中,过表达sgcR1将C-1027的产量提升至野生菌株的2–3倍,而在敲除sgcR的基础上过表达sgcR1将C-1027产量提升至37.5 mg/L,为野生菌株的7倍[83-84]。博来霉素(Bleomycin) 是属于糖肽类的广谱抗肿瘤抗生素,常用于治疗子宫癌和转移性睾丸癌等恶性肿瘤的联合化疗[85]。商用Blenoxane是博来霉素A2(36) 和B2(37) 以2︰1的比例组成的混合物,不易引起骨髓抑制或心脏毒性,可单独使用或与长春碱和顺式二氯铂联合使用[86]。由于博来霉素结构复杂,其生产依靠链霉菌发酵产生。2019年,Chen等通过敲除轮枝链霉菌Streptomyces verticillus中编码ArsR/SmtB家族调控因子的基因blmR,使得36和37的产量都得到了一定程度的提升。其中,37的产量较野生菌株提升了34%,达到56.66 mg/L (表 2)[87]。在棒状链霉菌Streptomyces clavuligerus OR中,同时过表达ccaR和编码LysR类型调控因子的基因claR将克拉维酸(38) 的产量提升了43%,达到了5.66 g/L[88]。
| Regulator | Strains | Antibiotics | Modea | Positionb | Yield (Fold/(mg/L)) | References |
| Tcs7 | S. tsukubaensis | Tacrolimus (26) | + | N | 1.4/204.1 | [65] |
| BlmR | S. verticillus | Bleomycin A2 (36) Bleomycin B2 (37) | – | M | – ~1.3/~56.7 | [87] |
| ClaR | S. clavuligerus | Clavulanic acid (38) | + | N | ~1.1/~4 360 | [88] |
| TlsB | S. anulatus | Telomestatin (39) | + | C | – | [89] |
| AcyB2 | S. thermotolerans | Carbomycins (40) | + | N | – | [90] |
| CtcS | S. aureofaciens | Chlortetracycline (15) | + | M | 1.2/– | [91] |
| AniF | S. hygrospinosus | Anisomycin (41) | + | C | 1.2/– | [35] |
| HcdR2 | S. mobaraensis | Herbicidin F (42) | + | C | 20/– | [92] |
| HmtA | S. hygroscopicus | Himastatin (43) | – | N | 9.9/~19 | [93] |
| IlaA | S. atratus | Ilamycins (44) | – | N | 3/~11.8 | [94] |
| DsaA | S. coelicolor | Desotamides (45) | + | M | 2.4/~30.6 | [95] |
| Bsm42 | S. spiramyceticus | Spiramycin (46) | + | N | – | [96] |
| Slinc377 | S. lincolnensis | Lincomycin (47) | – | M | ~2.4/~300 | [97] |
| Slinc191 | + | M | ~1.1/~187 | |||
| Slinc348 | + | N | ~7/~700 | |||
| CanR1 | S. albus | Candicidin (32) | + | N | 3.5/– | [98] |
| LnmO | S. atroolivaceus | Leinamycin (48) Leinamycin E1 (49) | + | C | ~3/1.6 ~4/84.00 |
[99] |
| LmbU | S. lincolnensis | Lincomycin (47) | + | M | 5/– | [100] |
| aThe following modes of regulation are indicated: +, activator; -, repressor. b Position of DNA-binding domain in the regulatory proteins are indicated: C, C-terminal; N, N-terminal; M, central. | ||||||
分别过表达tlsB、acyB2、ctcS、aniF、hcdR2以及hmtA调控基因,使得telomestatin (39)、carbomycins (40)、chlortetracycline (15)、anisomycin (41)、herbicidin F (42) 和himastatin (43) 的产量得到提升。其中,42的产量提升至原始菌株的20倍,而43产量提升到约19.02 mg/L,为原始菌株的9.9倍[35, 89-93]。Ilamycins (44) 属于一类结构新颖的环肽,具有较强的抗结核分枝杆菌的活性,是潜在的抗结核药物。He等在暗黑链霉菌中敲除ilaA,将44的产量在野生菌株的基础上提升了3倍,约为11.76 mg/L[94]。
在细菌中,ArsR类型调控因子通常为途径特异性负调控蛋白,但是链霉菌中几乎没有关于ArsR型调控因子的报道。Ding等将desotamides (45) 的生物合成基因簇在天蓝色链霉菌中异源表达,发现过表达编码ArsR家族调控因子的dsaA后,45的产量较出发菌株提升了2.4倍,说明dsaA是45生物合成基因簇中的途径特异性正调控基因[95]。在螺旋链霉菌Streptomyces spiramyceticus中过表达spiramycin (46) 生物合成基因簇中bsm42调控基因,提升了46的产量[96]。在林肯链霉菌Streptomyces lincolnensis中过表达编码ArsR/SmtB家族调控因子的slinc191和编码XRE家族调控因子的slinc348,将林克霉素(Lincomycin,47) 的产量分别提升至约187 mg/L和799 mg/L[97]。
人们对链霉菌途径特异性调控因子的研究不仅有助于链霉菌来源天然产物产量的提升,也有助于发现新颖的调控因子。例如,CanR1调控因子可以调控candicidin (32) 的生物合成,是在链霉菌中发现的第一个与次级代谢相关的XRE类型调控因子。Tian等发现CanR1通过特异性结合fscR1的启动子区,可以正向调控fscR1的表达。过表达canR1可以将32的产量提升至原始菌株的3.5倍[98]。雷纳霉素(leinamycin,48) 于1989年首次从链霉菌S. atroolivaceus S-140中发现,具有独特的1, 3-二氧-1, 2-二硫代烷结构。雷纳霉素E1 (Leinamycin E1,49) 是lnmE缺失突变菌株中发现的48的生物合成前体。化合物48和49均具有优异的抗肿瘤活性[101-102]。笔者发现雷纳霉素基因簇中的LnmO属于Crp/Fnr类型调控因子,但lnmO的缺失只是影响了48的产生,其菌体并无明显形态变化。在S. atroolivaceus野生菌株和lnmE缺失突变菌中分别过表达lnmO,可以将48和49的产量分别提高近3和4倍[99]。因此,LnmO是首个发现的Crp/Fnr类型途径特异性调控因子。Hou等发现在林可霉素(47) 的生物合成基因簇中,过表达lmbU将47的产量提升了5倍。LmbU的C端包含一个未知的结构域,而N端则包含一个自抑制域(Auto-inhibitory domain,AID),抑制LmbU的DNA结合活性。因此,LmbU属于一种新的调控因子,其DNA结合域通过HTH与DNA结合[103]。
3 总结与展望链霉菌产生的天然产物是小分子药物和药物先导化合物的重要来源。链霉菌来源天然产物的合成,在很大程度上受到其途径特异性调控因子的影响。因此,对这些调控因子功能的研究为提高链霉菌来源天然产物的产量奠定了基础(图 5)。据不完全统计,从2015年到2020年10月,研究人员通过多种手段对61个多种类型的调控因子进行了表征,其中TetR类型的有13个,包括7个正调控因子和6个负调控因子;LuxR类型的调控因子PAS-LuxR (12个) 和LAL (4个) 都属于正调控因子,而SARP (13个) 类型的调控因子也绝大多数属于正调控因子(图 5A)。通常通过对正调控基因的过表达,或敲除负调控基因,较大幅度提高了对应天然产物的产量,其中大部分提高幅度在1–5倍,包括5个工业菌株。过去的6年里,每年都有多篇关于链霉菌途径特异性调控因子的报道。根据通讯作者国籍进行统计,发现我国的研究人员的贡献占了主要部分(78%),说明我国对链霉菌途径特异性调控因子的研究颇具国际影响力。
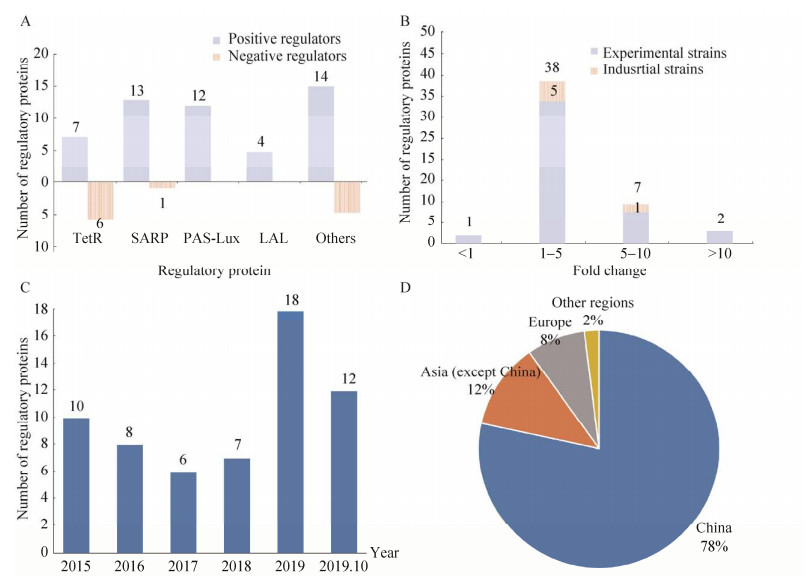
|
| 图 5 2015–2020年从链霉菌中发现的途径特异性调控因子的分析. (A) 不同家族、不同作用方式的调控蛋白数量;(B) 引起目标代谢产物产量不同幅度变化的调控因子数量;(C) 不同年份调控因子的发现数量;(D) 不同区域和国家对调控因子发现数量的贡献. Fig. 5 Analyses of pathway-specific regulators discovered from Streptomyces in 2015-2020. (A) Number of regulators belonging to different families. (B) Number of regulators triggering different fold changes of natural products. (C) Number of regulators discovered in different years. (D) Contribution to the discovery of regulators by major regions/ countries. |
| |
通过全局性和途径特异性调控因子的共同作用,链霉菌来源天然产物的生物合成严格受到级联调控;而部分生物合成基因簇内的调控基因,不仅控制着簇内基因的表达,同时对其他生物合成基因簇的表达也存在一定的影响,这形成了复杂的调控网络[104]。例如,geldanamycin和elaiophylin的生物合成通过gdmRIII关联起来,gdmRIII不仅在geldanamycin的生物合成中起正向调控作用,而且可以负调控elaiophylin的生物合成;在tetramycin和nystatin A1生物合成过程中,tetramycin簇内的调控基因ttmRIV同时调控这两个化合物的生物合成。这些调控基因对链霉菌来源天然产物生物合成的精细调控机制的阐释,将有助于理解天然产物在链霉菌生理和进化中的作用[105]。例如,candicidin生物合成基因簇里的4个调控基因fscR1–fscR4同为途径特异性正向调控基因,然而相对表达水平却存在较大差异,fscR1的表达水平比fscR4高出100倍左右,说明fscR1在促进candicidin生物合成中的主导地位。此外,这4个调控基因广泛分布在链霉菌多烯类天然产物的生物合成基因簇中,对这些天然产物及其调控网络的研究将有助于人们探索其他多烯天然产物的复杂调控机制。
对调控基因的深入研究,将更好地指导人们在合适的宿主中进行产量提升。如操纵调控基因sgcR和sgc1成功提升了C-1027产量,在此指导下,笔者后续发现的C-1027高产菌株中也能利用类似的策略实现产量提升[106]。比较不同菌株中C-1027的生物合成调控机制,为研究基因簇内的调控网络以及宿主的遗传背景提供了合适的平台。这将有助于阐明每个宿主的不同调控系统以及这些系统如何与相同的生物合成基因簇相互作用,从而为选择合适的异源宿主来实现产量提升提供指导[107]。在一些模式菌株(如天蓝色链霉菌、变铅青链霉菌等) 中进行异源表达也是构建高产菌株的有效方法。这是因为异源表达有效降低了亲本菌株中的代谢背景,并规避了复杂调控网络。在异源宿主中通过合理地对调控基因的操控以及启动子重组等策略,将大幅度促进目标代谢物产量的提升,或激活沉默的天然产物生物合成基因簇[108]。例如,通过在异源宿主中过表达调控基因dsaA提升了desotamides的产量[95],而过表达nonG[46]或srcmRI[60]分别激活了nocardamine或chromomycins的生物合成。在发酵工业中对途径特异性调控因子的研究,有助于提升工业链霉菌来源的天然产物产量,因而具有巨大的经济价值。例如,在已通过遗传育种发现的高产菌株龟裂链霉菌中过表达途径特异性调控基因otcR,将oxytetracycline的产量提升至出发菌株的6.49倍[52]。类似的是,过表达ccaR和claR进一步提高了高产工业菌株克拉维酸的产量,达到文献报道的最高水平[88]。
最后,由于大多数链霉菌来源的天然产物生物合成基因簇在典型实验室培养条件下是沉默的,对途径特异性调控基因的遗传操作是激活沉默基因簇的非常有效的手段。随着源于极端生态环境链霉菌资源的开发,通过链霉菌基因组测序揭示的大量新颖天然产物生物合成基因簇及对调控基因与其调控机制的深入研究,以及合成生物学技术在链霉菌来源的天然产物生物合成及调控研究中的广泛应用,相信将会有更多的具有优异生物活性的天然产物药物从被誉为“抗生素工厂”的链霉菌中发现。
| [1] |
Clardy J, Fischbach MA, Walsh CT. New antibiotics from bacterial natural products. Nat Biotechnol, 2006, 24(12): 1541-1550. DOI:10.1038/nbt1266
|
| [2] |
Shen B. A new golden age of natural products drug discovery. Cell, 2015, 163(6): 1297-1300. DOI:10.1016/j.cell.2015.11.031
|
| [3] |
Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod, 2020, 83(3): 770-803. DOI:10.1021/acs.jnatprod.9b01285
|
| [4] |
Bentley SD, Chater KF, Cerdeño-Tárraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 2002, 417(6885): 141-147. DOI:10.1038/417141a
|
| [5] |
Omura S, Ikeda H, Ishikawa J, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA, 2001, 98(21): 12215-12220. DOI:10.1073/pnas.211433198
|
| [6] |
Kalkreuter E, Pan GH, Cepeda AJ, et al. Targeting bacterial genomes for natural product discovery. Trends Pharmacol Sci, 2020, 41(1): 13-26. DOI:10.1016/j.tips.2019.11.002
|
| [7] |
Steele AD, Teijaro CN, Yang D, et al. Leveraging a large microbial strain collection for natural product discovery. J Biol Chem, 2019, 294(45): 16567-16576. DOI:10.1074/jbc.REV119.006514
|
| [8] |
Baltz RH. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol, 2016, 43(2/3): 343-370.
|
| [9] |
Kim HU, Charusanti P, Lee SY, et al. Metabolic engineering with systems biology tools to optimize production of prokaryotic secondary metabolites. Nat Prod Rep, 2016, 33(8): 933-941. DOI:10.1039/C6NP00019C
|
| [10] |
Smanski MJ, Peterson RM, Rajski SR, et al. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob Agents Chemother, 2009, 53(4): 1299-1304. DOI:10.1128/AAC.01358-08
|
| [11] |
Yu ZG, Smanski MJ, Peterson RM, et al. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org Lett, 2010, 12(8): 1744-1747. DOI:10.1021/ol100342m
|
| [12] |
Murakami T, Burian J, Yanai K, et al. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc Natl Acad Sci USA, 2011, 108(38): 16020-16025. DOI:10.1073/pnas.1108124108
|
| [13] |
Lu CY, Zhang XJ, Jiang M, et al. enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng, 2016, 35: 129-137. DOI:10.1016/j.ymben.2016.02.012
|
| [14] |
Wang WS, Li SS, Li ZL, et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat Biotechnol, 2020, 38(1): 76-83. DOI:10.1038/s41587-019-0335-4
|
| [15] |
Wang S, Deng H. The X-factor: enhanced β-oxidation on intracellular triacylglycerols enabling overproduction of polyketide drug-like molecules in microorganisms. Synth Syst Biotechnol, 2020, 5(1): 19-20. DOI:10.1016/j.synbio.2020.01.005
|
| [16] |
Wang B, Zhao HM. Unleashing the power of energy storage: engineering β-oxidation pathways for polyketide production. Synth Syst Biotechnol, 2020, 5(1): 21-22. DOI:10.1016/j.synbio.2020.01.002
|
| [17] |
Romero-Rodríguez A, Robledo-Casados I, Sánchez S. An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta, 2015, 1849(8): 1017-1039. DOI:10.1016/j.bbagrm.2015.06.007
|
| [18] |
Wei JH, He L, Niu GQ. Regulation of antibiotic biosynthesis in actinomycetes: perspectives and challenges. Synth Syst Biotechnol, 2018, 3(4): 229-235. DOI:10.1016/j.synbio.2018.10.005
|
| [19] |
Xia HY, Li XF, Li ZQ, et al. The application of regulatory cascades in Streptomyces: yield enhancement and metabolite mining. Front Microbiol, 2020, 11: 406-420. DOI:10.3389/fmicb.2020.00406
|
| [20] |
Xia HY, Zhan XQ, Mao XM, et al. The regulatory cascades of antibiotic production in Streptomyces. World J Microbiol Biotechnol, 2020, 36(1): 13-22. DOI:10.1007/s11274-019-2789-4
|
| [21] |
Martín JF, Liras P. Cascades and networks of regulatory genes that control antibiotic biosynthesis. Subcell Biochem, 2012, 64: 115-138.
|
| [22] |
Van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep, 2011, 28(7): 1311-1333. DOI:10.1039/c1np00003a
|
| [23] |
Martin JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol, 2004, 186(16): 5197-5201. DOI:10.1128/JB.186.16.5197-5201.2004
|
| [24] |
Zhu H, Sandiford SK, Van Wezel GP. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol, 2014, 41(2): 371-386. DOI:10.1007/s10295-013-1309-z
|
| [25] |
Liu G, Chater KF, Chandra G, et al. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev, 2013, 77(1): 112-143. DOI:10.1128/MMBR.00054-12
|
| [26] |
Scherlach K, Hertweck C. Triggering cryptic natural productbiosynthesis in microorganisms. Org Biomol Chem, 2009, 7(9): 1753-1760. DOI:10.1039/b821578b
|
| [27] |
Ochi K. Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J Antibiot (Tokyo), 2017, 70(1): 25-40. DOI:10.1038/ja.2016.82
|
| [28] |
Ramos JL, Martínez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev, 2005, 69(2): 326-356. DOI:10.1128/MMBR.69.2.326-356.2005
|
| [29] |
Ahn SK, Tahlan K, Yu Z, et al. Investigation of transcription repression and small-molecule responsiveness by TetR-like transcription factors using a heterologous Escherichia coli-based assay. J Bacteriol, 2007, 189(18): 6655-6664. DOI:10.1128/JB.00717-07
|
| [30] |
Yu Z, Reichheld SE, Savchenko A, et al. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol, 2010, 400(4): 847-864. DOI:10.1016/j.jmb.2010.05.062
|
| [31] |
Cuthbertson L, Nodwell JR. The TetR family of regulators. Microbiol Mol Biol Rev, 2013, 77(3): 440-475. DOI:10.1128/MMBR.00018-13
|
| [32] |
Liang JL, Nie Y, Wang MX, et al. Regulation of alkane degradation pathway by a TetR family repressor via an autoregulation positive feedback mechanism in a Gram-positive Dietzia bacterium. Mol Microbiol, 2016, 99(2): 338-359. DOI:10.1111/mmi.13232
|
| [33] |
Baral B, Akhgari A, Metsä-Ketelä M. Activation of microbial secondary metabolic pathways: avenues and challenges. Synth Syst Biotechnol, 2018, 3(3): 163-178. DOI:10.1016/j.synbio.2018.09.001
|
| [34] |
Santos CL, Correia-Neves M, Moradas-Ferreira P, et al. A walk into the LuxR regulators of Actinobacteria: phylogenomic distribution and functional diversity. PLoS ONE, 2012, 7(10): e46758. DOI:10.1371/journal.pone.0046758
|
| [35] |
Shen JF, Kong LX, Li Y, et al. A LuxR family transcriptional regulator AniF promotes the production of anisomycin and its derivatives in Streptomyces hygrospinosus var. beijingensis. Synth Syst Biotechnol, 2019, 4(1): 40-48. DOI:10.1016/j.synbio.2018.12.004
|
| [36] |
Barreales EG, Vicente CM, De Pedro A, et al. Promoter engineering reveals the importance of heptameric direct repeats for DNA binding by Streptomyces antibiotic regulatory protein-large ATP-binding regulator of the LuxR family (SARP-LAL) regulators in Streptomyces natalensis. Appl Environ Microbiol, 2018, 84(10): e00246-18.
|
| [37] |
Bhukya H, Jana AK, Sengupta N, et al. Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence. J Struct Biol, 2017, 198(2): 134-146. DOI:10.1016/j.jsb.2017.03.006
|
| [38] |
Baltz RH. Biosynthesis and genetic engineering of lipopeptide antibiotics related to daptomycin. Curr Top Med Chem, 2008, 8(8): 618-638. DOI:10.2174/156802608784221497
|
| [39] |
Yu GH, Jia XQ, Wen JP, et al. Strain improvement of Streptomyces roseosporus for daptomycin production by rational screening of He-Ne laser and NTG induced mutants and kinetic modeling. Appl Biochem Biotechnol, 2011, 163(6): 729-743. DOI:10.1007/s12010-010-9078-x
|
| [40] |
Yuan PH, Zhou RC, Chen XP, et al. DepR1, a TetR family transcriptional regulator, positively regulates daptomycin production in an industrial producer, Streptomyces roseosporus SW0702. Appl Environ Microbiol, 2016, 82(6): 1898-1905. DOI:10.1128/AEM.03002-15
|
| [41] |
Zhao HY, Wang L, Wan D, et al. Characterization of the aurantimycin biosynthetic gene cluster and enhancing its production by manipulating two pathway-specific activators in Streptomyces aurantiacus JA 4570. Microb Cell Fact, 2016, 15: 160-172. DOI:10.1186/s12934-016-0559-7
|
| [42] |
Li Y, Li JJ, Tian ZH, et al. Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J Biol Chem, 2016, 291(10): 5406-5417. DOI:10.1074/jbc.M115.695874
|
| [43] |
Jiang MX, Yin M, Wu SH, et al. GdmRⅢ, a TetR family transcriptional regulator, controls geldanamycin and elaiophylin biosynthesis in Streptomyces autolyticus CGMCC0516. Sci Rep, 2017, 7: 4803. DOI:10.1038/s41598-017-05073-x
|
| [44] |
Wei KK, Wu YJ, Li L, et al. MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol, 2018, 102(20): 8841-8853. DOI:10.1007/s00253-018-9280-2
|
| [45] |
Fidan O, Yan RM, Zhu D, et al. Improved production of antifungal angucycline Sch47554 by manipulating three regulatory genes in Streptomyces sp. SCC-2136. Biotechnol Appl Biochem, 2019, 66(4): 517-526. DOI:10.1002/bab.1748
|
| [46] |
Park W, Woo JK, Shin J, et al. NonG, a constituent of the nonactin biosynthetic gene cluster, regulates nocardamine synthesis in Streptomyces albus J1074. Biochem Biophys Res Commun, 2017, 490(3): 664-669. DOI:10.1016/j.bbrc.2017.06.098
|
| [47] |
Yoo YJ, Hwang JY, Shin HL, et al. Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production. J Ind Microbiol Biotechnol, 2015, 42(1): 125-135. DOI:10.1007/s10295-014-1546-9
|
| [48] |
Mast Y, Guezguez J, Handel F, et al. A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl Environ Microbiol, 2015, 81(19): 6621-6636. DOI:10.1128/AEM.00728-15
|
| [49] |
Gou LX, Han TS, Wang XX, et al. A novel TetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front Microbiol, 2017, 8: 2371-2381. DOI:10.3389/fmicb.2017.02371
|
| [50] |
Ye SH, Braña AF, González-Sabín J, et al. New insights into the biosynthesis pathway of polyketide alkaloid argimycins P in Streptomyces argillaceus. Front Microbiol, 2018, 9: 252-264. DOI:10.3389/fmicb.2018.00252
|
| [51] |
Lyu M, Cheng YQ, Han X, et al. AccR, a TetR family transcriptional repressor, coordinates short-chain acyl coenzyme a homeostasis in Streptomyces avermitilis. Appl Environ Microbiol, 2020, 86(12): e00508-00520.
|
| [52] |
Yin SL, Wang WS, Wang XF, et al. Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb Cell Fact, 2015, 14: 46-58. DOI:10.1186/s12934-015-0231-7
|
| [53] |
Chen YW, Liu XC, Lv FX, et al. Characterization of three regulatory genes involved in enduracidin biosynthesis and improvement of enduracidin production in Streptomyces fungicidicus. J Appl Microbiol, 2019, 127(6): 1698-1705. DOI:10.1111/jam.14417
|
| [54] |
Yang K, Qi LH, Zhang M, et al. The SARP family regulator Txn9 and two-component response regulator Txn11 are key activators for trioxacarcin biosynthesis in Streptomyces bottropensis. Curr Microbiol, 2015, 71(4): 458-464. DOI:10.1007/s00284-015-0868-9
|
| [55] |
刘佳, 朱涛, 王鹏飞, 等. 金霉素生物合成基因簇中调控基因ctcB的功能. 微生物学报, 2016, 56(9): 1486-1495. Liu J, Zhu T, Wang PF, et al. Function of Streptomyces antibiotic regulatory proteins family transcriptional regulator ctcB in the biosynthetic cluster of chlortetracycline. Acta Microbiol Sin, 2016, 56(9): 1486-1495 (in Chinese). |
| [56] |
Wu XR, Jin L, Zhang H, et al. Identification of truncated form of NosP as a transcription factor to regulate the biosynthesis of nosiheptide. FASEB J, 2018, 32(1): 453-465. DOI:10.1096/fj.201700556r
|
| [57] |
Li Y, Kong LX, Shen JF, et al. Characterization of the positive SARP family regulator PieR for improving piericidin A1 production in Streptomyces piomogeues var. Hangzhouwanensis. Synth Syst Biotechnol, 2019, 4(1): 16-24. DOI:10.1016/j.synbio.2018.12.002
|
| [58] |
Li Y, Zhang JH, Zheng JZ, et al. Co-expression of a SARP family activator ChlF2 and a type Ⅱ thioesterase ChlK led to high production of chlorothricin in Streptomyces antibioticus DSM 40725. Front Bioeng Biotechnol, 2020, 8: 1013-1025. DOI:10.3389/fbioe.2020.01013
|
| [59] |
Goh F, Zhang MM, Lim TR, et al. Identification and engineering of 32 membered antifungal macrolactone notonesomycins. Microb Cell Fact, 2020, 19(1): 71-84. DOI:10.1186/s12934-020-01328-x
|
| [60] |
Sun L, Zeng J, Cui PW, et al. Manipulation of two regulatory genes for efficient production of chromomycins in Streptomyces reseiscleroticus. J Biol Eng, 2018, 12: 9-20. DOI:10.1186/s13036-018-0103-x
|
| [61] |
Koomsiri W, Inahashi Y, Leetanasaksakul K, et al. Sarpeptins A and B, lipopeptides produced by Streptomyces sp. KO-7888 overexpressing a specific SARP regulator. J Nat Prod, 2019, 82(8): 2144-2151. DOI:10.1021/acs.jnatprod.9b00074
|
| [62] |
Yang ZJ, Wei X, He JQ, et al. Characterization of the noncanonical regulatory and transporter genes in atratumycin biosynthesis and production in a heterologous host. Mar Drugs, 2019, 17(10): 560-571. DOI:10.3390/md17100560
|
| [63] |
Cui H, Ni XP, Shao W, et al. Functional manipulations of the tetramycin positive regulatory gene ttmRIV to enhance the production of tetramycin A and nystatin A1 in Streptomyces ahygroscopicus. J Ind Microbiol Biotechnol, 2015, 42(9): 1273-1282. DOI:10.1007/s10295-015-1660-3
|
| [64] |
Zhang YY, He HR, Liu H, et al. Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb Cell Fact, 2016, 15(1): 152-169. DOI:10.1186/s12934-016-0552-1
|
| [65] |
Zhang XS, Luo HD, Tao Y, et al. FkbN and Tcs7 are pathway-specific regulators of the FK506 biosynthetic gene cluster in Streptomyces tsukubaensis L19. J Ind Microbiol Biotechnol, 2016, 43(12): 1693-1703. DOI:10.1007/s10295-016-1849-0
|
| [66] |
Zhu ZH, Li H, Yu P, et al. SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl Microbiol Biotechnol, 2017, 101(4): 1547-1557. DOI:10.1007/s00253-016-7918-5
|
| [67] |
Guan HY, Li Y, Zheng JZ, et al. Important role of a LAL regulator StaR in the staurosporine biosynthesis and high-production of Streptomyces fradiae CGMCC 4.576. Sci China Life Sci, 2019, 62(12): 1638-1654. DOI:10.1007/s11427-019-1597-6
|
| [68] |
Martín JF, Ramos A, Liras P. Regulation of geldanamycin biosynthesis by cluster-situated transcription factors and the master regulator PhoP. Antibiotics, 2019, 8(3): 87-103. DOI:10.3390/antibiotics8030087
|
| [69] |
Li C, He HR, Wang JB, et al. Characterization of a LAL-type regulator NemR in nemadectin biosynthesis and its application for increasing nemadectin production in Streptomyces cyaneogriseus. Sci China Life Sci, 2019, 62(3): 394-405. DOI:10.1007/s11427-018-9442-9
|
| [70] |
Zhang Y, Chen H, Wang P, et al. Identification of the regulon FkbN for ascomycin biosynthesis and its interspecies conservation analysis as LAL family regulator. Biochem Eng J, 2019, 151: 12-24.
|
| [71] |
Xu J, Song ZQ, Xu XH, et al. ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol, 2019, 103(17): 7071-7084. DOI:10.1007/s00253-019-09959-w
|
| [72] |
Zhu YP, Xu WH, Zhang J, et al. A hierarchical network of four regulatory genes controlling production of the polyene antibiotic candicidin in Streptomyces sp. strain FR-008. Appl Environ Microbiol, 2020, 86(9): e00055-e00020.
|
| [73] |
Cheng ZL, Bown L, Tahlan K, et al. Regulation of coronafacoyl phytotoxin production by the PAS-LuxR family regulator CfaR in the common scab pathogen Streptomyces scabies. PLoS ONE, 2015, 10(3): e0122450. DOI:10.1371/journal.pone.0122450
|
| [74] |
Yang J, Xu DX, Yu WC, et al. Regulation of aureofuscin production by the PAS-LuxR family regulator AurJ3M. Enzyme Microb Technol, 2020, 137: 109532-109546. DOI:10.1016/j.enzmictec.2020.109532
|
| [75] |
Kurniawan YN, Kitani S, Maeda A, et al. Differential contributions of two SARP family regulatory genes to indigoidine biosynthesis in Streptomyces lavendulae FRI-5. Appl Microbiol Biotechnol, 2014, 98(23): 9713-9721. DOI:10.1007/s00253-014-5988-9
|
| [76] |
Olano C, Lombó F, Méndez C, et al. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng, 2008, 10(5): 281-292. DOI:10.1016/j.ymben.2008.07.001
|
| [77] |
Santos-Beneit F, Rodríguez-García A, Martín JF. Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiol Res, 2012, 167(9): 532-535. DOI:10.1016/j.micres.2012.02.010
|
| [78] |
Ma DX, Wang C, Chen H, et al. Manipulating the expression of SARP family regulator BulZ and its target gene product to increase tacrolimus production. Appl Microbiol Biotechnol, 2018, 102(11): 4887-4900. DOI:10.1007/s00253-018-8979-4
|
| [79] |
Santos-Aberturas J, Vicente CM, Guerra SM, et al. Molecular control of polyene macrolide biosynthesis: direct binding of the regulator PimM to eight promoters of pimaricin genes and identification of binding boxes. J Biol Chem, 2011, 286(11): 9150-9161. DOI:10.1074/jbc.M110.182428
|
| [80] |
Antón N, Santos-Aberturas J, Mendes MV, et al. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology, 2007, 153(Pt 9): 3174-3183.
|
| [81] |
Santos-Aberturas J, Payero TD, Vicente CM, et al. Functional conservation of PAS-LuxR transcriptional regulators in polyene macrolide biosynthesis. Metab Eng, 2011, 13(6): 756-767. DOI:10.1016/j.ymben.2011.09.011
|
| [82] |
Chen YH, Smanski MJ, Shen B. Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol, 2010, 86(1): 19-25. DOI:10.1007/s00253-009-2428-3
|
| [83] |
Chen YH, Yin M, Horsman GP, et al. Manipulation of pathway regulation in Streptomyces globisporus for overproduction of the enediyne antitumor antibiotic C-1027. J Antibiot (Tokyo), 2010, 63(8): 482-485. DOI:10.1038/ja.2010.55
|
| [84] |
Chen YH, Yin M, Horsman GP, et al. Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus. J Nat Prod, 2011, 74(3): 420-424. DOI:10.1021/np100825y
|
| [85] |
Du LC, Sánchez C, Chen M, et al. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem Biol, 2000, 7(8): 623-642. DOI:10.1016/S1074-5521(00)00011-9
|
| [86] |
Latta VD, Cecchettini A, Del Ry S, et al. Bleomycin in the setting of lung fibrosis induction: from biological mechanisms to counteractions. Pharmacol Res, 2015, 97: 122-130. DOI:10.1016/j.phrs.2015.04.012
|
| [87] |
Chen H, Wang JH, Cui JQ, et al. Negative regulation of bleomycins biosynthesis by ArsR/SmtB family repressor BlmR in Streptomyces verticillus. Appl Microbiol Biotechnol, 2019, 103(16): 6629-6644. DOI:10.1007/s00253-019-09923-8
|
| [88] |
Cho HS, Jo JC, Shin CH, et al. Improved production of clavulanic acid by reverse engineering and overexpression of the regulatory genes in an industrial Streptomyces clavuligerus strain. J Ind Microbiol Biotechnol, 2019, 46(8): 1205-1215. DOI:10.1007/s10295-019-02196-0
|
| [89] |
Amagai K, Ikeda H, Hashimoto J, et al. Identification of a gene cluster for telomestatin biosynthesis and heterologous expression using a specific promoter in a clean host. Sci Rep, 2017, 7(1): 3382-3340. DOI:10.1038/s41598-017-03308-5
|
| [90] |
Zhong JJ, Lu ZL, Dai JL, et al. Identification of two regulatory genes involved in carbomycin biosynthesis in Streptomyces thermotolerans. Arch Microbiol, 2017, 199(7): 1023-1033. DOI:10.1007/s00203-017-1376-z
|
| [91] |
Kong LX, Liu J, Zheng XQ, et al. CtcS, a MarR family regulator, regulates chlortetracycline biosynthesis. BMC Microbiol, 2019, 19(1): 279-290. DOI:10.1186/s12866-019-1670-9
|
| [92] |
Shi YY, Gu RJ, Li YH, et al. Exploring novel herbicidin analogues by transcriptional regulator overexpression and MS/MS molecular networking. Microb Cell Fact, 2019, 18(1): 175-192. DOI:10.1186/s12934-019-1225-7
|
| [93] |
Xie YC, Li QL, Qin XJ, et al. Enhancement of himastatin bioproduction via inactivation of atypical repressors in Streptomyces hygroscopicus. Metab Eng Commun, 2019, 8: e00084. DOI:10.1016/j.mec.2018.e00084
|
| [94] |
He JQ, Wei X, Yang ZJ, et al. Characterization of regulatory and transporter genes in the biosynthesis of anti-tuberculosis ilamycins and production in a heterologous host. Mar Drugs, 2020, 18(4): 216-227. DOI:10.3390/md18040216
|
| [95] |
Ding WJ, Dong YL, Ju JH, et al. The roles of genes associated with regulation, transportation, and macrocyclization in desotamide biosynthesis in Streptomyces scopuliridis SCSIO ZJ46. Appl Microbiol Biotechnol, 2020, 104(6): 2603-2610. DOI:10.1007/s00253-020-10414-4
|
| [96] |
Dai JL, Wang YG, Liu JJ, et al. The regulatory genes involved in spiramycin and bitespiramycin biosynthesis. Microbiol Res, 2020, 240: 126532-126541. DOI:10.1016/j.micres.2020.126532
|
| [97] |
Wang RD, Kong FJ, Wu HZ, et al. Complete genome sequence of high-yield strain S. lincolnensis B48 and identification of crucial mutations contributing to lincomycin overproduction. Synth Syst Biotechnol, 2020, 5(2): 37-48. DOI:10.1016/j.synbio.2020.03.001
|
| [98] |
Tian J, Ye LX, Yang YL, et al. Transposon-based screen identifies a XRE family regulator crucial for candicidin biosynthesis in Streptomyces albus J1074. Sci China Life Sci, 2020, 63(9): 1-4. DOI:10.1007/s11427-019-1582-5
|
| [99] |
Huang Y, Yang D, Pan GH, et al. Characterization of LnmO as a pathway-specific Crp/Fnr-type positive regulator for leinamycin biosynthesis in Streptomyces atroolivaceus and its application for titer improvement. Appl Microbiol Biotechnol, 2016, 100(24): 10555-10562. DOI:10.1007/s00253-016-7864-2
|
| [100] |
Hou BB, Lin YW, Wu HZ, et al. The novel transcriptional regulator LmbU promotes lincomycin biosynthesis through regulating expression of its target genes in Streptomyces lincolnensis. J Bacteriol, 2018, 200(2): e00447-00417.
|
| [101] |
Pan GH, Xu ZR, Guo ZK, et al. Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc Natl Acad Sci USA, 2017, 114(52): E11131-E11140. DOI:10.1073/pnas.1716245115
|
| [102] |
Huang SX, Yun BS, Ma M, et al. Leinamycin E1 acting as an anticancer prodrug activated by reactive oxygen species. Proc Natl Acad Sci USA, 2015, 112(27): 8278-8283. DOI:10.1073/pnas.1506761112
|
| [103] |
Hou BB, Zhu XY, Kang YJ, et al. LmbU, a cluster-situated regulator for lincomycin, consists of a DNA-binding domain, an auto-inhibitory domain, and forms homodimer. Front Microbiol, 2019, 10: 989-1002. DOI:10.3389/fmicb.2019.00989
|
| [104] |
Bednarz B, Kotowska M, Pawlik KJ. Multi-level regulation of coelimycin synthesis in Streptomyces coelicolor A3(2). Appl Microbiol Biotechnol, 2019, 103(16): 6423-6434. DOI:10.1007/s00253-019-09975-w
|
| [105] |
Castro-Melchor M, Charaniya S, Karypis G, et al. Genome-wide inference of regulatory networks in Streptomyces coelicolor. BMC Genomics, 2010, 11: 578-594. DOI:10.1186/1471-2164-11-578
|
| [106] |
Yan XH, Hindra, Ge HM, et al. Discovery of alternative producers of the enediyne antitumor antibiotic C-1027 with high titers. J Nat Prod, 2018, 81(3): 594-599. DOI:10.1021/acs.jnatprod.7b01013
|
| [107] |
Teijaro CN, Adhikari A, Shen B. Challenges and opportunities for natural product discovery, production, and engineering in native producers versus heterologous hosts. J Ind Microbiol Biotechnol, 2019, 46(3/4): 433-444. DOI:10.1007/s10295-018-2094-5
|
| [108] |
Chaudhary AK, Dhakal D, Sohng JK. An insight into the "-omics" based engineering of streptomycetes for secondary metabolite overproduction. Biomed Res Int, 2013, 2013: 968518-968533.
|
 2021, Vol. 37
2021, Vol. 37