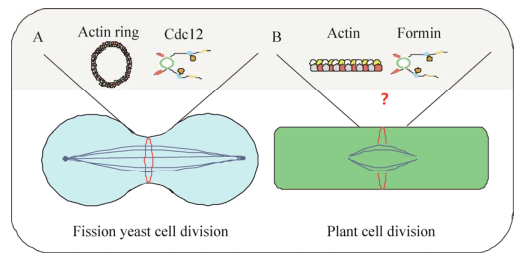
| [1] | |
|
| [2] | |
|
| [3] |
Blanchoin L, Staiger CJ. Plant formins: diverse isoforms and unique molecular mechanism. Biochim Biophys Acta (BBA) - Mol Cell Res, 2010, 1803(2): 201-206. DOI:10.1016/j.bbamcr.2008.09.015
|
|
| [4] |
Cvrčková F, Novotný M, Pícková D, et al. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics, 2004, 5: 44. DOI:10.1186/1471-2164-5-44
|
|
| [5] |
Vidali L, Van Gisbergen PAC, Guérin C, et al. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA, 2009, 106(32): 13341-13346. DOI:10.1073/pnas.0901170106
|
|
| [6] |
Xu YW, Moseley JB, Sagot I, et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell, 2004, 116(5): 711-723. DOI:10.1016/S0092-8674(04)00210-7
|
|
| [7] |
Shimada A, Nyitrai M, Vetter IR, et al. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell, 2004, 13(4): 511-522. DOI:10.1016/S1097-2765(04)00059-0
|
|
| [8] |
Kitayama C, Uyeda TQP. ForC, a novel type of formin family protein lacking an FH1 domain, is involved in multicellular development in Dictyostelium discoideum . J Cell Sci, 2003, 116(4): 711-723. DOI:10.1242/jcs.00265
|
|
| [9] |
Grunt M, Zárský V, Cvrčková F. Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evolut Biol, 2008, 8: 115. DOI:10.1186/1471-2148-8-115
|
|
| [10] | |
|
| [11] |
Zeng YF, Xiao YS, Liu Y, et al. Formin-like 3 regulates RhoC/FAK pathway and actin assembly to promote cell invasion in colorectal carcinoma. World J Gastroenterol, 2018, 24(34): 3884-3897. DOI:10.3748/wjg.v24.i34.3884
|
|
| [12] | |
|
| [13] |
Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell, 2005, 16(1): 1-13. DOI:10.1091/mbc.e04-07-0565
|
|
| [14] |
Brandizzi F, Barlowe C. Organization of the ER-golgi interface for membrane traffic control. Nat Rev Mol Cell Biol, 2013, 14(6): 382-392. DOI:10.1038/nrm3588
|
|
| [15] | |
|
| [16] |
Huang J, Kim CM, Xuan YH, et al. Formin homology 1 ( OsFH1) regulates root-hair elongation in rice ( Oryza sativa). Planta, 2013, 237(5): 1337-1239. DOI:10.1007/s00425-013-1840-1
|
|
| [17] |
Lan YX, Liu XN, Fu Y, et al. Arabidopsis class Ⅰ formins control membrane-originated actin polymerization at pollen tube tips. PLoS Genet, 2018, 14(11): e1007789. DOI:10.1371/journal.pgen.1007789
|
|
| [18] |
Van Gisbergen PAC, Bezanilla M. Plant formins: membrane anchors for actin polymerization. Trends Cell Biol, 2013, 23(5): 227-233. DOI:10.1016/j.tcb.2012.12.001
|
|
| [19] |
Petersen J, Nielsen O, Egel R, et al. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol, 1998, 141(5): 1217-1228. DOI:10.1083/jcb.141.5.1217
|
|
| [20] |
Le SM, Yu M, Bershadsky A, et al. Mechanical regulation of formin-dependent actin polymerization. Semin Cell Dev Biol, 2020, 102: 73-80. DOI:10.1016/j.semcdb.2019.11.016
|
|
| [21] | |
|
| [22] |
Pruyne D. Revisiting the phylogeny of the animal formins: two new subtypes, relationships with multiple wing hairs proteins, and a lost human formin. PLoS ONE, 2016, 11(10): e0164067. DOI:10.1371/journal.pone.0164067
|
|
| [23] |
Lu J, Meng WY, Poy F, et al. Structure of the FH2 domain of daam1: implications for formin regulation of actin assembly. J Mol Biol, 2007, 369(5): 1258-1269. DOI:10.1016/j.jmb.2007.04.002
|
|
| [24] |
Cvrčková F, Oulehlová D, Žárský V. Formins: linking cytoskeleton and endomembranes in plant cells. Int J Mol Sci, 2014, 16(1): 1-18. DOI:10.3390/ijms16010001
|
|
| [25] |
Vizcarra CL, Bor B, Quinlan ME. The role of formin tails in actin nucleation, processive elongation, and filament bundling. J Biol Chem, 2014, 289(44): 30602-30613. DOI:10.1074/jbc.M114.588368
|
|
| [26] |
Pernier J, Shekhar S, Jegou A, et al. Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching, and motility. Dev Cell, 2016, 36(2): 201-214. DOI:10.1016/j.devcel.2015.12.024
|
|
| [27] |
Deeks MJ, Fendrych M, Smertenko A, et al. The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J Cell Sci, 2010, 123(8): 1209-1215. DOI:10.1242/jcs.065557
|
|
| [28] |
Van Gisbergen PAC, Li M, Wu SZ, et al. Class Ⅱ formin targeting to the cell cortex by binding PI(3, 5)P 2 is essential for polarized growth. J Cell Biol, 2012, 198(2): 235-250. DOI:10.1083/jcb.201112085
|
|
| [29] |
Cifrová P, Oulehlová D, Kollárová E, et al. Division of labor between two actin nucleators-the formin FH1 and the ARP2/3 complex-in Arabidopsis epidermal cell morphogenesis. Front Plant Ence, 2020, 11: 148. DOI:10.3389/fpls.2020.00148
|
|
| [30] |
Liu C, Zhang Y, Ren HY. Actin polymerization mediated by AtFH5 directs the polarity establishment and vesicle trafficking for pollen germination in Arabidopsis . Mol Plant, 2018, 11(11): 1389-1399. DOI:10.1016/j.molp.2018.09.004
|
|
| [31] |
Yi KX, Guo CQ, Chen D, et al. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis . Plant Physiol, 2005, 138(2): 1071-1082. DOI:10.1104/pp.104.055665
|
|
| [32] |
Oulehlová D, Kollárová E, Cifrová P, et al. Arabidopsis class Ⅰ Formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol, 2019, 60(8): 1855-1870. DOI:10.1093/pcp/pcz102
|
|
| [33] |
Zhang Z, Zhang Y, Tan HX, et al. Rice morphology determinant encodes the type Ⅱ formin FH5 and regulates rice morphogenesis. Plant Cell, 2011, 23(2): 681-700. DOI:10.1105/tpc.110.081349
|
|
| [34] | |
|
| [35] | |
|
| [36] |
Takatsuka H, Higaki T, Umeda M. Actin reorganization triggers rapid cell elongation in roots. Plant Physiol, 2018, 178(3): 1130-1141. DOI:10.1104/pp.18.00557
|
|
| [37] |
Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci, 2003, 116(13): 2603-2611. DOI:10.1242/jcs.00611
|
|
| [38] | |
|
| [39] | |
|
| [40] |
Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol, 2001, 11(21): 1656-1665. DOI:10.1016/S0960-9822(01)00525-5
|
|
| [41] | |
|
| [42] |
Lammers M, Meyer S, Kühlmann D, et al. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem, 2008, 283(50): 35236-35246. DOI:10.1074/jbc.M805634200
|
|
| [43] |
Chen H, Kuo CC, Kang H, et al. Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol Biol Cell, 2012, 23(19): 3814-3826. DOI:10.1091/mbc.e12-05-0400
|
|
| [44] |
Wallar BJ, Deward AD, Resau JH, et al. RhoB and the mammalian diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res, 2007, 313: 560-571. DOI:10.1016/j.yexcr.2006.10.033
|
|
| [45] |
Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev, 2006, 70(3): 605-645. DOI:10.1128/MMBR.00013-06
|
|
| [46] |
Dominguez R. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem Sci, 2016, 41(6): 478-490. DOI:10.1016/j.tibs.2016.03.004
|
|
| [47] |
Skruber K, Warp PV, Shklyarov R, et al. Arp2/3 and Mena/VASP require profilin 1 for actin network assembly at the leading edge. Curr Biol, 2020, 30(14): 2651-2664. DOI:10.1016/j.cub.2020.04.085
|
|
| [48] |
Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton, 2009, 66(8): 606-617. DOI:10.1002/cm.20379
|
|
| [49] |
Courtemanche N, Pollard TD. Determinants of formin homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem, 2012, 287(10): 7812-7820. DOI:10.1074/jbc.M111.322958
|
|
| [50] |
Billault-Chaumartin IB, Martin SG. Capping protein insulates Arp2/3-assembled actin patches from formins. Curr Biol, 2019, 29(19): 3165-3176. DOI:10.1016/j.cub.2019.07.088
|
|
| [51] | |
|
| [52] |
Favery B, Chelysheva LA, Lebris M, et al. Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell, 2004, 16(9): 2529-2540. DOI:10.1105/tpc.104.024372
|
|
| [53] |
Zheng YY, Xin HB, Lin JX, et al. An Arabidopsis class Ⅱ formin, AtFH19, nucleates actin assembly, binds to the barbed end of actin filaments, and antagonizes the effect of AtFH1 on actin dynamics. J Integr Plant Biol, 2012, 54(10): 800-813. DOI:10.1111/j.1744-7909.2012.01160.x
|
|
| [54] |
Kollárová E, Forero AB, Stillerová L, et al. Arabidopsis class Ⅱ formins AtFH13 and AtFH14 can form heterodimers but exhibit distinct patterns of cellular localization. Int J Mol Sci, 2020, 21(1): 348. DOI:10.3390/ijms21010348
|
|
| [55] |
Cheung AY, Wu HM. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell, 2004, 16(1): 257-269. DOI:10.1105/tpc.016550
|
|
| [56] |
Ingouff M, Gerald JNF, Guérin C, et al. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol, 2005, 7(4): 374-380. DOI:10.1038/ncb1238
|
|
| [57] |
Michelot A, Guérin C, Huang SJ, et al. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell, 2005, 17(8): 2296-2313. DOI:10.1105/tpc.105.030908
|
|
| [58] | |
|
| [59] |
Yi PS, Goshima G. Rho of plants GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during Physcomitrella patens branching. Curr Biol, 2020, 30(14): 2860-2868. DOI:10.1016/j.cub.2020.05.022
|
|
| [60] |
Oda Y, Fukuda H. Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell, 2013, 25(11): 4439-4450. DOI:10.1105/tpc.113.117853
|
|
| [61] | |
|
| [62] | |
|
| [63] |
Liu T, Wang YW, Wang YB, et al. Multifaceted regulation of PTEN subcellular distributions and biological functions. Cancers, 2019, 11(9): 1247. DOI:10.3390/cancers11091247
|
|
| [64] |
Fragoso R, Barata JT. Kinases, tails and more: regulation of PTEN function by phosphorylation. Methods, 2015, 77-78: 75-81. DOI:10.1016/j.ymeth.2014.10.015
|
|
| [65] |
Wang JJ, Zhang Y, Wu J, et al. AtFH16, an Arabidopsis type Ⅱ formin, binds and bundles both microfilaments and microtubules, and preferentially binds to microtubules. J Integrat Plant Biol, 2013, 55(11): 1002-1015. DOI:10.1111/jipb.12089
|
|
| [66] |
Yang N, Ray SD, Krafts K. Cell proliferation// Wexler P, Ed. Encyclopedia of toxicology. 3rd ed. Oxford: Academic Press, 2014: 761-765.
|
|
| [67] | |
|
| [68] |
Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature, 2010, 463(7280): 485-492. DOI:10.1038/nature08908
|
|
| [69] |
Huisman SM, Brunner D. Cell polarity in fission yeast: a matter of confining, positioning, and switching growth zones. Semin Cell Dev Biol, 2011, 22(8): 799-805. DOI:10.1016/j.semcdb.2011.07.013
|
|
| [70] |
Akram Z, Ahmed I, Mack H, et al. Yeast as a model to understand actin-mediated cellular functions in mammals-illustrated with four actin cytoskeleton proteins. Cells, 2020, 9(3): 672. DOI:10.3390/cells9030672
|
|
| [71] | |
|
| [72] |
Snaith HA, Samejima I, Sawin KE. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J, 2005, 24(21): 3690-3699. DOI:10.1038/sj.emboj.7600838
|
|
| [73] | |
|
| [74] |
Vidali L, Bezanilla M. Physcomitrella patens : a model for tip cell growth and differentiation. Curr Opin Plant Biol, 2012, 15(6): 625-631. DOI:10.1016/j.pbi.2012.09.008
|
|
| [75] |
Yang WB, Ren SL, Zhang XM, et al. BENT UPPERMOST INTERNODE1 encodes the class Ⅱ formin FH5 crucial for actin organization and rice development. Plant Cell, 2011, 23(2): 661-680. DOI:10.1105/tpc.110.081802
|
|
| [76] |
Rosero A, Žárský V, Cvrčková F. AtFH1 formin mutation affects actin filament and microtubule dynamics in Arabidopsis thaliana . J Exp Bot, 2012, 64(2): 585-597.
|
|
| [77] |
Dong YQ, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J Cell Biol, 2003, 161(6): 1081-1092. DOI:10.1083/jcb.200212040
|
|
| [78] |
Xue XH, Guo CQ, Du F, et al. AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol Plant, 2011, 4(2): 264-278. DOI:10.1093/mp/ssq085
|
|
| [79] | |
|
| [80] | |
|
 2021, Vol. 37
2021, Vol. 37