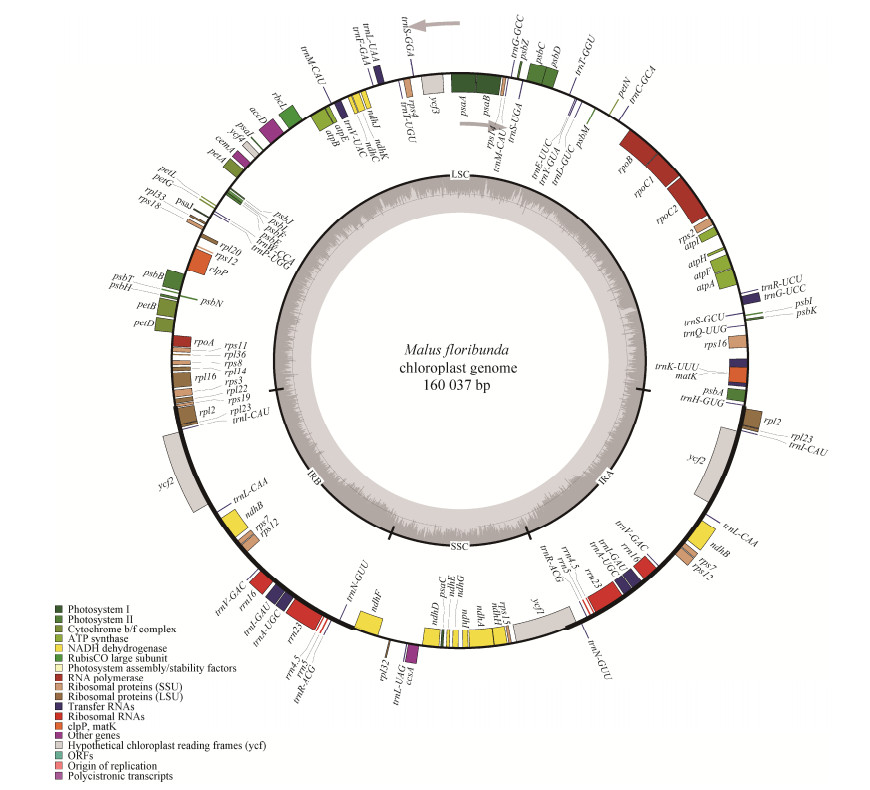
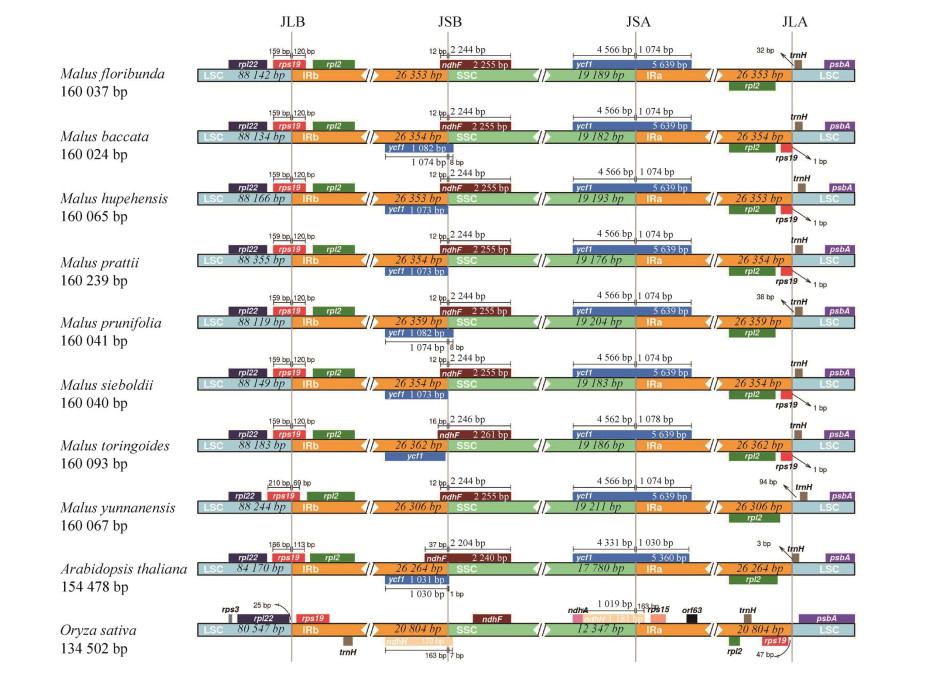
| [1] |
周婷, 沈星诚, 周道建, 等. 海棠品种分类研究进展. 园艺学报, 2018, 45(2): 380-396. Zhou T, Shen XC, Zhou DJ, et al. Advances in the classification of crabapple cultivars. Acta Hortic Sin, 2018, 45(2): 380-396 (in Chinese). DOI:10.16420/j.issn.0513-353x.2017-0140
|
|
| [2] |
李育农. 苹果属植物种质资源研究. 北京: 中国农业出版社, 2001.
Li YN. Researches of Germplasm Resources of Malus Mill. Beijing: Chinese Agriculture Press, 2001 (in Chinese).
|
|
| [3] |
姜文龙, 范俊俊, 张丹丹, 等. 观赏海棠不同叶位色彩特征及特异种质挖掘. 园艺学报, 2017, 44(6): 1135-1144. Jiang WL, Fan JJ, Zhang DD, et al. Research on the characteristics of different leaf positions of crabapple's leaf color and elite germplasm excavation. Acta Hortic Sin, 2017, 44(6): 1135-1144 (in Chinese). DOI:10.16420/j.issn.0513-353x.2016-0774
|
|
| [4] |
郑杨, 曲晓玲, 郭翎, 等. 观赏海棠资源谱系分析及育种研究进展. 山东农业大学学报(自然科学版), 2008, 39(1): 152-160. Zheng Y, Qu XL, Guo L, et al. Advances on ornamental crabapple resources. J Shandong Agric Univ (Nat Sci Ed), 2008, 39(1): 152-160 (in Chinese). DOI:10.3969/j.issn.1000-2324.2008.01.033
|
|
| [5] |
聂继云, 吕德国, 李静, 等. 22种苹果种质资源果实类黄酮分析. 中国农业科学, 2010, 43(21): 4455-4462. Nie JY, Lü DG, Li J, et al. A preliminary study on the flavonoids in fruits of 22 apple germplasm resources. Sci Agric Sin, 2010, 43(21): 4455-4462 (in Chinese). DOI:10.3864/j.issn.0578-1752.2010.21.015
|
|
| [6] |
孙希生, 丛佩华, 薛光荣. 德国苹果抗性育种概况. 中国果树, 2002(4): 57-58. Sun XS, Cong PH, Xue GR. General situation of resistance breeding of apple in Germany. China Fruits, 2002(4): 57-58 (in Chinese). DOI:10.3969/j.issn.1000-8047.2002.04.033
|
|
| [7] |
Papp D, Singh J, Gadoury D, et al. New North American isolates of Venturia inaequalis can overcome apple scab resistance of Malus floribunda 821. Plant Dis, 2020, 104(3): 649-655. DOI:10.1094/PDIS-10-19-2082-RE
|
|
| [8] |
闫忠业, 娄汉平, 田颖辉. 苹果抗病虫育种研究概况. 中国果树, 2005(6): 48-49, 58. Yan ZY, Lou HP, Tian YH. The survey overview of apple breeding for resistance to disease and insect. China Fruits, 2005(6): 48-49, 58 (in Chinese). DOI:10.3969/j.issn.1000-8047.2005.06.026
|
|
| [9] |
高源, 王大江, 王昆, 等. 苹果属15个种的叶绿体DNA变异与遗传分化. 果树学报, 2021, 38(1): 1-12. Gao Y, Wang DJ, Wang K, et al. Genetic divergence and population structure of chloroplast DNA in fifteen species of Malus Mill. J Fruit Sci, 2021, 38(1): 1-12 (in Chinese). DOI:10.13925/j.cnki.gsxb.20200205
|
|
| [10] |
李慧峰. 泰沂山区苹果属植物系统学研究[D]. 沈阳: 沈阳农业大学, 2012.
Li HF. Studies on the taxonomy of the genus Malus Mill. (Rosaceae)of Taiyi-mountains[D]. Shenyang: Shenyang Agricultural University, 2012 (in Chinese).
|
|
| [11] |
李亚楠. 苹果属(Malus Mill. )种间关系的分子系统学研究[D]. 哈尔滨: 东北林业大学, 2021.
Li YN. Molecular systematics study on interspecific relationship of Malus Mill[D]. Harbin: Northeast Forestry University, 2021 (in Chinese).
|
|
| [12] |
Wang S, Cai XR, Zhang B, et al. Characterization of the complete chloroplast genome sequence of Malus toringoides (Rosaceae). Mitochondrial DNA B Resour, 2020, 5(3): 2603-2604. DOI:10.1080/23802359.2020.1781572
|
|
| [13] |
Li YN, Liu YL, Wu P, et al. The complete chloroplast genome sequence of Malus toringoides (Rosaceae). Mitochondrial DNA B Resour, 2020, 5(3): 2787-2789. DOI:10.1080/23802359.2020.1780977
|
|
| [14] |
Jin JJ, Yu WB, Yang JB, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol, 2020, 21(1): 241. DOI:10.1186/s13059-020-02154-5
|
|
| [15] |
Wick RR, Schultz MB, Zobel J, et al. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 2015, 31(20): 3350-3352. DOI:10.1093/bioinformatics/btv383
|
|
| [16] |
Qu XJ, Moore MJ, Li DZ, et al. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 2019, 15: 50. DOI:10.1186/s13007-019-0435-7
|
|
| [17] |
Liu C, Shi LC, Zhu YJ, et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics, 2012, 13: 715. DOI:10.1186/1471-2164-13-715
|
|
| [18] |
Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res, 2019, 47(W1): W59-W64.
|
|
| [19] | |
|
| [20] |
Kurtz S, Choudhuri JV, Ohlebusch E, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res, 2001, 29(22): 4633-4642. DOI:10.1093/nar/29.22.4633
|
|
| [21] |
Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res, 1987, 15(3): 1281-1295. DOI:10.1093/nar/15.3.1281
|
|
| [22] |
Frazer KA, Pachter L, Poliakov A, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res, 2004, 32(Web Server issue): W273-W279.
|
|
| [23] |
Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics, 2018, 34(17): 3030-3031. DOI:10.1093/bioinformatics/bty220
|
|
| [24] |
Bi GQ, Mao YX, Xing QK, et al. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics, 2018, 110(1): 18-22. DOI:10.1016/j.ygeno.2017.08.001
|
|
| [25] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096
|
|
| [26] |
Liu BB, Hong DY, Zhou SL, et al. Phylogenomic analyses of the Photinia complex support the recognition of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Rosaceae). J Syst Evol, 2019, 57(6): 678-694. DOI:10.1111/jse.12542
|
|
| [27] |
Zhang X, Rong CX, Qin L, et al. Complete chloroplast genome sequence of Malus hupehensis: genome structure, comparative analysis, and phylogenetic relationships. Molecules, 2018, 23(11): 2917. DOI:10.3390/molecules23112917
|
|
| [28] |
Bao L, Li K, Liu Z, et al. Characterization of the complete chloroplast genome of the Chinese crabapple Malus prunifolia (Rosales: Rosaceae: Maloideae). Conserv Genet Resour, 2016, 8(3): 227-229. DOI:10.1007/s12686-016-0540-0
|
|
| [29] |
左瑞华, 蒋平, 孙传伯, 等. 基于全基因组从头测序技术盐肤木叶绿体基因组的测序分析. 生物工程学报, 2020, 36(4): 772-781. Zuo RH, Jiang P, Sun CB, et al. Analysis of the chloroplast genome characteristics of Rhus chinensis by de novo sequencing. Chin J Biotech, 2020, 36(4): 772-781 (in Chinese).
|
|
| [30] | |
|
| [31] |
李连星, 彭劲谕, 王大玮, 等. 长爪栘[木衣]叶绿体基因组特征系统发育及密码子偏好性分析. 生物工程学报, 2022, 38(1): 328-342. Li LX, Peng JY, Wang DW, et al. Chloroplast genome phylogeny and codon preference of Docynia longiunguis. Chin J Biotech, 2022, 38(1): 328-342 (in Chinese).
|
|
| [32] | |
|
| [33] |
周晓君, 张凯, 彭正锋, 等. 矮牡丹与芍药属其他5个种叶绿体基因组特征的比较. 林业科学, 2020, 56(4): 82-88. Zhou XJ, Zhang K, Peng ZF, et al. Comparative analysis of chloroplast genome characteristics between Paeonia jishanensis and other five species of Paeonia. Sci Silvae Sin, 2020, 56(4): 82-88 (in Chinese).
|
|
| [34] |
朱斌, 甘晨晨, 王洪程. 球花石斛( Dendrobium thyrsiflorum)叶绿体基因组特征及亲缘关系解析. 生物技术通报, 2021, 37(5): 38-47. Zhu B, Gan CC, Wang HC. Characteristics of the complete chloroplast genome of Dendrobium thyrsiflorum and its phylogenetic relationship analysis. Biotechnol Bull, 2021, 37(5): 38-47 (in Chinese).
|
|
| [35] |
郑祎, 张卉, 王钦美, 等. 大花君子兰叶绿体基因组及其特征. 园艺学报, 2020, 47(12): 2439-2450. Zheng Y, Zhang H, Wang QM, et al. Complete chloroplast genome sequence of Clivia miniata and its characteristics. Acta Hortic Sin, 2020, 47(12): 2439-2450 (in Chinese).
|
|
| [36] |
李泳潭, 张军, 黄亚丽, 等. 杜梨叶绿体基因组分析. 园艺学报, 2020, 47(6): 1021-1032. Li YT, Zhang J, Huang YL, et al. Analysis of chloroplast genome of Pyrus betulaefolia. Acta Hortic Sin, 2020, 47(6): 1021-1032 (in Chinese).
|
|
| [37] |
高源, 王大江, 王昆, 等. 新疆野苹果叶绿体DNA变异与遗传进化分析. 植物遗传资源学报, 2020, 21(3): 579-587. Gao Y, Wang DJ, Wang K, et al. Chloroplast DNA variation and genetic evolution of Malus sieversii (Ledeb.) M. Roem. J Plant Genet Resour, 2020, 21(3): 579-587 (in Chinese).
|
|
| [38] |
高源, 王大江, 王昆, 等. 基于叶绿体DNA变异的山荆子种质遗传多样性和系统演化. 中国农业科学, 2020, 53(3): 600-611. Gao Y, Wang DJ, Wang K, et al. Genetic diversity and phylogenetics of Malus baccata (L. ) borkh revealed by chloroplast DNA variation. Sci Agric Sin, 2020, 53(3): 600-611 (in Chinese).
|
|
| [39] |
高源, 王大江, 王昆, 等. 基于叶绿体DNA分析的楸子种质遗传多样性研究. 园艺学报, 2020, 47(5): 853-863. Gao Y, Wang DJ, Wang K, et al. Genetic diversity of Malus prunifolia germplasms based on chloroplast DNA analysis. Acta Hortic Sin, 2020, 47(5): 853-863 (in Chinese).
|
|
| [40] |
李裕华, 任永康, 赵兴华, 等. 禾本科主要农作物叶绿体基因组研究进展. 生物技术通报, 2020, 36(11): 112-121. Li YH, Ren YK, Zhao XH, et al. Research progress on chloroplast genome of major gramineous crops. Biotechnol Bull, 2020, 36(11): 112-121 (in Chinese).
|
|
 2022, Vol. 38
2022, Vol. 38