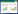中国科学院微生物研究所、中国微生物学会主办
文章信息
- 杨小涵, 伍国强, 魏明, 王北辰
- YANG Xiaohan, WU Guoqiang, WEI Ming, WANG Beichen
- 甜菜BvHAK基因家族全基因组鉴定及其在盐处理下的表达分析
- Genome-wide identification of BvHAK gene family in sugar beet (Beta vulgaris) and their expression analysis under salt treatments
- 生物工程学报, 2022, 38(10): 3773-3789
- Chinese Journal of Biotechnology, 2022, 38(10): 3773-3789
- 10.13345/j.cjb.220257
-
文章历史
- Received: April 5, 2022
- Accepted: July 18, 2022
盐胁迫是制约全球农作物生长和产量的主要环境因素之一[1]。全球盐碱地面积多达9.5亿hm2,约占陆地总面积的7%[2]。我国是世界上土壤盐碱化最为严重的国家之一,盐渍化土地面积多达0.99亿hm2,约占全国土地面积的1/10,主要分布在西北、华北、东北及沿海地区[3]。绝大多数农作物、尤其是甜土植物(glycophytes) 对盐分很敏感[4]。土壤中高浓度盐分会扰乱植物对N、P、K等元素的吸收,产生离子毒害及渗透胁迫并诱发氧化胁迫,从而抑制生长、甚至导致作物的绝产[5-6]。当植物遭受盐胁迫时,介质中的Na+将通过根系非选择性阳离子通道(non-selective cation channels, NSCCs) 和高亲和性K+转运蛋白(high-affinity K+ transporters, HKTs) 进入细胞,高浓度Na+会紊乱离子的稳态平衡[7-8]。因此,维持或重建离子平衡对植物应答盐胁迫至关重要。
植物主要通过大量吸收K+和外排Na+来维持体内离子稳态平衡,以抵御盐胁迫[9]。HAK/KUP/KT (high-affinity K+ transporter/K+ uptake permease/K+ transporter) 家族是植物中最大的K+转运蛋白家族之一,其在盐胁迫下通过电化学梯度介导高亲和性K+吸收,从而维持植物体内K+/Na+稳态平衡,增强植物的耐盐性[10]。在拟南芥(Arabidopsis thaliana) 中,AtKUP6和AtKUP11受盐胁迫的诱导并上调表达[11]。研究表明,过量表达水稻(Oryza sativa) OsHAK5使转基因烟草植株体内K+积累显著增加,从而提高植株的耐盐性[12]。目前,HAK基因家族已在许多物种中被鉴定,包括大豆(Glycine max)[13]、葡萄(Vitis vinifera)[14]、橡胶树(Hevea brasliensis)[15]、马铃薯(Solanum tuberosum)[16]、香蕉(Musa acuminata)[17]、油菜(Brassica napus)[18]、小麦(Triticum aestivum)[19]等。然而,在糖料作物中尚未见HAK基因家族研究的报道。
在我国北方地区广为种植的糖料作物甜菜(Beta vulgaris L.) 为藜科(Chenopodiaceae) 甜菜属(Beta) 二年生草本植物,属于典型的嗜盐作物,具有很强的耐盐碱性和出色的耐贫瘠性,是盐碱地轮作和改良的重要作物[20]。研究表明,添加适量NaCl (15−100 mmol/L) 不仅能够显著促进甜菜幼苗生长,而且还能明显减缓渗透胁迫对其造成的不利影响;即使在未添加NaCl的条件下,渗透胁迫后的甜菜叶中仍能够维持K+/Na+稳态平衡[21]。本课题组前期基于甜菜基因组数据库[22],采用生物信息学手段,挖掘和鉴定出BvNHX[23]、BvWKRY[24]、BvSnRK2[25]和BvCIPK[26]基因家族成员,并阐明了它们在甜菜响应盐胁迫中的作用机制。然而,目前尚未见对甜菜BvHAK基因家族成员鉴定及其盐处理下表达分析的报道。
鉴于此,本研究对甜菜BvHAK基因家族进行全基因组鉴定及表达模式分析,探究BvHAK在甜菜响应盐胁迫中的作用,以期为农作物抗逆性遗传改良提供基因资源和理论依据。
1 材料与方法 1.1 材料供试甜菜(B. vulgaris L.) 品种为“甘糖7号”,种子购自甘肃省威武三农种业科技有限公司。挑选籽粒饱满的种子,播种至装满蛭石的育苗盘(5 cm×5 cm×5 cm; 32孔),每孔播3粒种子。用蒸馏水浇灌,待种子萌发后,使用改良的1/2 Hoagland营养液浇灌。待出苗两周后,进行间苗,每孔留1株生长均匀一致的幼苗。幼苗培养条件:温度25 ℃/20 ℃ (昼/夜),光周期16 h/8 h (昼/夜),光照强度550−600 μmol/(m2·s),空气相对湿度65%−75%。培养至4周龄的甜菜幼苗,分别用0 (对照)、50、100和150 mmol/L NaCl处理72 h后,地上部和根分开取样,3次生物学重复,经液氮速冻后置于–80 ℃冰箱保存备用[27]。
1.2 甜菜BvHAK基因家族成员的鉴定利用拟南芥基因组数据库TAIR (http://wwwarabidopsis.org) 获得13个AtHAK基因的氨基酸序列[28]。然后利用NCBI基因组数据库(http://www.ncbi.nlm.nih.gov) 和甜菜基因组数据库(http://bvseq.boku.ac.at/index.shtml),通过BLAST在线搜索BvHAK基因家族成员序列[29]。BvHAK预选蛋白的所有同源蛋白序列均满足期望值(E) < 10–40。
通过ExPASy (http://web.expasy.org/) 在线数据库计算BvHAK基因家族成员的等电点(isoelectric point, pI) 和分子量(molecular weight, MW)[30]。通过在线软件Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) 预测BvHAK基因家族成员的亚细胞定位[31]。采用TMHMM 3.0 (http://www.cbs.dtu.dk/serbices/TMHMM) 鉴定BvHAK基因家族编码蛋白的跨膜结构域(transmembrane domain, TM)[32]。
1.3 甜菜BvHAK染色体分布分析利用甜菜基因组数据库确定BvHAK基因家族成员在染色体上的定位,并通过MapInspect 1.0软件(http://mapinspect.software.informer.com/) 绘制BvHAK基因家族成员在染色体上的分布图[33]。
1.4 HAK家族系统进化关系及dN/dS分析为探究HAK基因家族进化及系统发育关系,采用MEGA X软件(http://www.megasoftware.net/) 对拟南芥、水稻、玉米(Zea mays)[34]、大麦(Hordeum vulgare)[35]、小麦[36]、白杨(Populus alba)、菠菜(Spinacia oleracea) 和甜菜等9个物种111个HAK基因家族蛋白序列进行系统进化分析,并通过邻接比对(neighbor-joining, NJ) 法以1 000重复算法构建系统进化树[37]。采用PAL2NAL在线软件(
利用在线工具MEME 5.4.1 (https://meme-suite.org/meme/) 预测BvHAK基因家族成员的蛋白质保守基序(motif),基序的最大数目为10,宽度设置为6−50。采用GSDS在线网站(http://gsds.cbi.pku.edu.cn/) 分析BvHAK基因家族成员的基因结构[39]。通过将编码区序列(coding domain sequence, CDS) 与相应基因组序列(http://bvseq.boku.ac.at) 进行比对,生成BvHAK基因家族成员的外显子/内含子结构图。
1.6 甜菜BvHAK家族蛋白三维结构和互作网络分析利用SWISS-MODEL在线软件(https://swissmodel.expasy.org/interactive) 预测了BvHAK蛋白的三维结构(three-dimensional structure, 3-D) 模型[40]。利用STRING在线数据库(http://string-db.org) 进行蛋白与蛋白相互作用预测。
1.7 甜菜BvHAK基因家族成员顺式作用元件和基因表达模式分析利用plantCARE在线软件(http://bioinformation.psb.ugent.be/webtools/plantcare/html) 预测BvHAK基因家族成员启动子中可能存在的各种顺式作用调控元件[41]。
采用UNIQ-10柱式Trizol总RNA抽提试剂盒(生工生物工程(上海) 股份有限公司) 对不同浓度NaCl处理的甜菜根和叶进行总RNA提取,利用cDNA第一链合成试剂盒(PrimeScriptTM RT Master Mix, TaKaRa) 合成cDNA第一链。采用Primer Premier 5.0和Oligo7.0软件设计用于qRT-PCR分析的基因特异性引物(表 1)[42]。按照荧光实时定量PCR试剂盒(TB GreenTM Premix Ex TaqTM Ⅱ, TaKaRa) 进行qRT-PCR反应[43]。BvACTIN作为内参基因。通过2–ΔΔCt方法进行计算BvHAK基因的相对表达水平。所有的结果均以3个生物学重复的平均值±SE表示,每个生物学重复由3个技术重复组成。采用SPSS 22.0软件分析显著性差异水平(P < 0.05),并用Excel 2010作图[44]。
| Gene name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
| BvHAK1 | CTACAAACACGAATTAATGGCTTCT | AAAGCCAACCGCAGAAGCAATAG |
| BvHAK2 | GCACAATGGATTTATCAACTCACCC | CAGAATACCAAAGAAAGAACACCAAA |
| BvHAK3 | AATGTAAGCAGTAACAATGAGCAAGA | GCGGAATGACCACCACCTACAT |
| BvHAK4 | AACAATAAGGAGAAATCATGGAGGAC | AACACATACAAAGGGGATATACTCA |
| BvHAK5 | TTAGCTTATCAGAGTCTTGGAATAGT | AAACAGTGAGGCTCCAAAAGATCAA |
| BvHAK6 | TGTGGGTGAAAAAATGGTAGGAAAAG | ACTCACTTCACTTCCATCTACCCA |
| BvHAK7 | TGTGCCCTTTTTGAGTGAGTCTATG | GCTTAAATCTCCATACACTACTCCT |
| BvHAK8 | GTGTCGGTATCGTTTATGGGGATT | CCCATTATCATCTGCGCTCAATAC |
| BvHAK9 | GGATGATGGCGAGGAGAGAGAAAG | ACCTATCTTCAATTCCGAAACTTCA |
| BvHAK10 | CTAACATGGATCTGGAAAACAACAA | AAAACAAAGGACAAAACCCCATAAAT |
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
在甜菜基因组中共鉴定到10个BvHAK基因家族成员,并根据其所对应的染色体上的位置信息,依此对鉴定的基因命名为BvHAK1− BvHAK10 (表 2)。序列分析表明,BvHAK基因家族成员的CDS范围为2 004−2 541 bp,编码氨基酸的个数在668−847 aa不等,平均值为778.3 aa;分子量为81.76−95.16 kDa,平均值为88.31 kDa。亚细胞定位预测结果显示,BvHAK1、-4、-6、-7、-9和-10等6个成员定位于液泡膜(vacuole, Vac),而BvHAK2、-3、-5和-8等4个成员定位于质膜(plasma membrane, PM) (表 2)。
| Gene name | Gene ID | Chr | Exon count | CDS (bp) | Protein length (aa) | pI | MW (kDa) | TM | Subcellular localization |
| BvHAK1 | Bv1_012350_gwcc | 1 | 9 | 2 418 | 806 | 7.30 | 89.87 | 14 | Vac |
| BvHAK2 | Bv2_038040_xuej | 2 | 10 | 2 199 | 733 | 8.96 | 81.76 | 12 | PM |
| BvHAK3 | Bv2_043210_nsir | 2 | 8 | 2 286 | 762 | 8.37 | 85.29 | 11 | PM |
| BvHAK4 | Bv2_030090_hets | 2 | 9 | 2 358 | 786 | 6.49 | 88.16 | 12 | Vac |
| BvHAK5 | Bv5_099680_qewf | 5 | 9 | 2 325 | 775 | 9.41 | 87.53 | 12 | PM |
| BvHAK6 | Bv6_149150_dnus | 6 | 9 | 2 523 | 841 | 7.19 | 93.59 | 13 | Vac |
| BvHAK7 | Bv6_149160_jraq | 6 | 8 | 2 334 | 778 | 7.59 | 87.46 | 13 | Vac |
| BvHAK8 | Bv6_146500_ndso | 6 | 9 | 2 349 | 783 | 8.76 | 87.38 | 12 | PM |
| BvHAK9 | Bv7_171890_ygwk | 7 | 10 | 2 541 | 847 | 5.38 | 95.16 | 12 | Vac |
| BvHAK10 | Bv_005060_xrpy | UN | 8 | 2 316 | 772 | 6.95 | 86.92 | 12 | Vac |
为进一步确定BvHAK基因间的结构差异,对BvHAK家族成员进行基因结构分析。如图 1A所示,10个BvHAK基因含有8−10个外显子和7−9个内含子。其中,BvHAK3、-5、-7和-10等4个成员各含有8个外显子和7个内含子;BvHAK1、-4、-6和-8等4个成员各含有9个外显子和8个内含子;BvHAK2和-9各含有10个外显子和9个内含子。另外,BvHAK基因家族成员外显子长度及内含子结构呈现出相对保守的特点(图 1A)。
|
| 图 1 甜菜BvHAK家族基因结构(A) 和染色体定位(B) 分析 Fig. 1 Analysis of the gene structure (A) and chromosome location (B) of BvHAK gene family members. |
| |
甜菜BvHAK基因家族染色体定位分析结果表明,10个家族成员中有9个成员能够精准定位在9条甜菜基因组的1、2、5、6、7号染色体上(图 1B)。1、5和7号染色体上各有1个成员,分别是BvHAK1、-5和-9;2号染色体上有3个成员,分别是BvHAK2、-3和-4;6号染色体上也分布3个成员,分别为BvHAK6、-7和-8。然而,BvHAK10没有定位在染色体上。
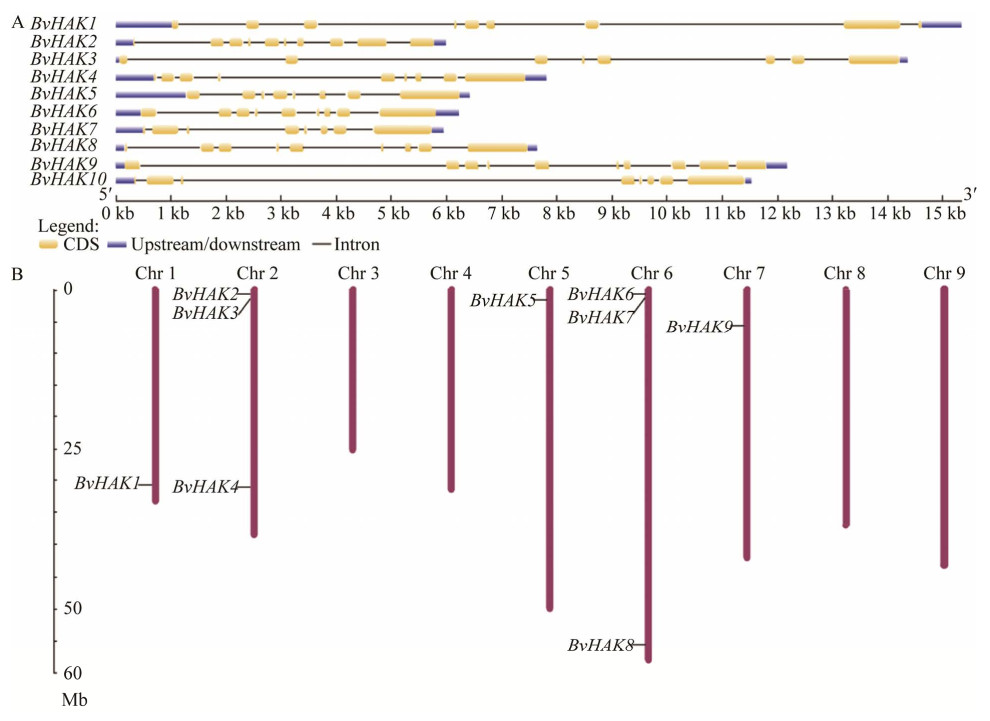
2.3 HAK基因家族系统进化树构建和dN/dS分析为解析甜菜与其他物种之间HAK基因家族成员的进化关系,对甜菜10个、拟南芥13个、菠菜12个、白杨10个、大麦6个、水稻27个、玉米21个、小麦8个、大麦4个共计111个HAK基因家族成员氨基酸序列进行系统发育树构建。结果表明,HAK基因家族成员分为5簇,分别为Ⅰ、Ⅱ、Ⅲ、Ⅴ和Ⅳ簇,其中Ⅱ簇成员可分为Ⅱa、Ⅱb和Ⅱc等3个亚簇(图 2)。甜菜BvHAK基因家族成员分布在前4簇中,分别为1个、6个、1个和2个成员,而第Ⅳ簇中没有BvHAK基因家族成员。其中,BvHAK3在第I簇;BvHAK5和BvHAK8在Ⅱa簇;BvHAK2在Ⅱb簇;BvHAK4、BvHAK7和BvHAK10在Ⅱc簇,BvHAK1在第Ⅲ簇;而BvHAK6和BvHAK9在第Ⅴ簇中。在同一簇中,甜菜BvHAK基因家族成员在进化树中与菠菜、拟南芥、白杨的亲缘关系距离更近,而与玉米、水稻、小麦、大麦等单子叶植物的进化关系较远(图 2)。
|
| 图 2 植物HKT基因家族系统发育树 Fig. 2 Phylogenetic tree of HKT family in plants. |
| |
在遗传学中,未导致氨基酸改变的核苷酸变异称为同义突变,反之则称为非同义突变。一般认为,同义突变不受自然选择,而非同义突变则受到自然选择作用。dN/dS表示异义替换(dN) 和同义替换(dS) 之间的比例。这个比例可以判断是否有选择压力作用于蛋白质编码基因。若dN/dS > 1,则认为有正选择效应。若dN/dS=1,则认为存在中性选择。若dN/dS < 1,则认为有纯化选择作用。如表 3所示,大多数BvHAKs (除BvHAK8和BvHAK10外) 与BvHAK3的dN/dS均大于1,表明BvHAK1、-2、-4、-5、-6、-7和-9与BvHAK3在进化上具有正选择作用。进一步采用PAML4.9软件对10个BvHAK基因家族的CDS序列进行选择压力分析。结果表明,BvHAK序列选择压力检测参数dN/dS小于1 (数据未显示),可见BvHAKs基因所承受的选择压力总体趋势为纯化选择。
| Gene1 | Gene2 | dS | dN | dN/dS |
| BvHAK1 | BvHAK3 | 1.046 5 | 17.897 8 | 17.102 4 |
| BvHAK2 | BvHAK3 | 2.134 6 | 6.860 2 | 3.213 8 |
| BvHAK4 | BvHAK3 | 1.486 3 | 11.868 7 | 7.985 5 |
| BvHAK5 | BvHAK3 | 0.101 1 | 10.004 9 | 99.000 0 |
| BvHAK6 | BvHAK3 | 1.880 2 | 7.450 0 | 3.962 3 |
| BvHAK7 | BvHAK3 | 3.283 3 | 3.476 8 | 1.058 9 |
| BvHAK8 | BvHAK3 | 12.977 9 | 4.446 8 | 0.342 6 |
| BvHAK9 | BvHAK3 | 0.150 3 | 14.878 | 99.000 0 |
| BvHAK10 | BvHAK3 | 5.106 3 | 4.478 8 | 0.877 1 |
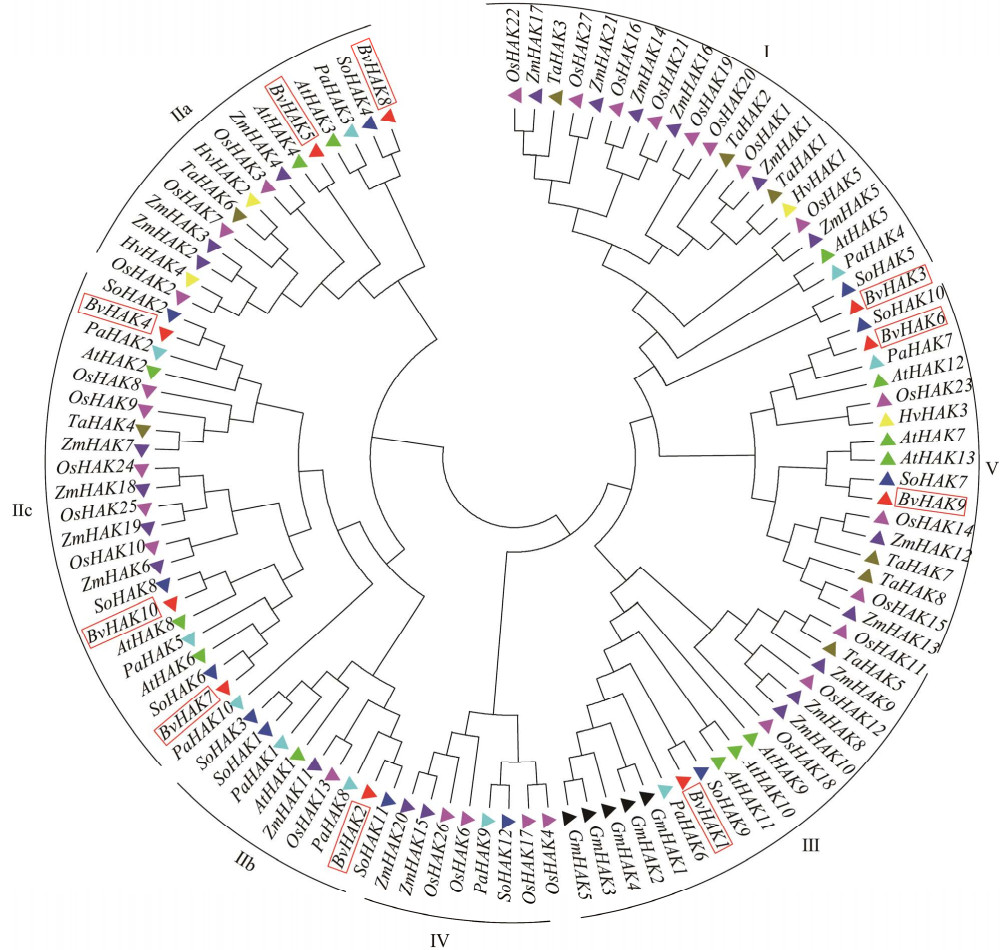
根据序列比对结果可以看出(图 3),所有BvHAK的TM2和TM3之间有一个长的胞质环(loop),此为高亲和性K+转运蛋白典型的结构特点。基于MEME分析设置10个基序,分析了甜菜BvHAK基因编码氨基酸序列,发现所有的BvHAK基因家族成员均含有motif1、motif2、motif3、motif4、motif5、motif6、motif7、motif8、motif9和motif10 (图 4)。另外,同一簇中,各个成员基序的位置基本上保持一致。由此可见,BvHAK基因家族具有高度保守的结构域。另外,BvHAK蛋白跨膜结构域的数量在11–14不等,等电点位于5.38−9.41之间,其中7个成员的pI大于7 (表 2),表明绝大多数BvHAK属于碱性蛋白。
|
| 图 3 甜菜BvHAK基因家族蛋白保守结构域氨基酸序列比对TM2和TM3分别代表第2和第3个跨膜区 Fig. 3 Amino acid sequence alignment of the protein conserved domains of BvHAK genes. TM2 and TM3 indicate the 2nd and 3rd transmembrane domain, respectively. |
| |
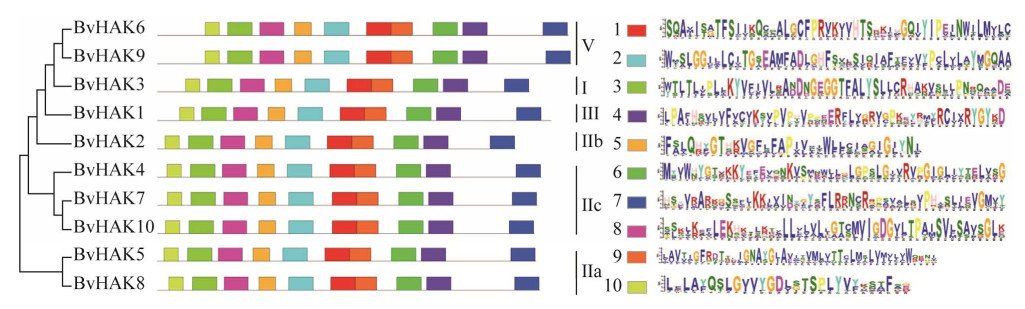
|
| 图 4 甜菜BvHAK基因家族蛋白保守基序分布及特征分析 Fig. 4 Distribution and characteristics of conserved motifs in BvHAK genes. |
| |
为了进一步研究BvHAK基因家族的蛋白质结构,采用SWISS-MODEL软件预测BvHAK蛋白3-D结构(图 5),所有蛋白的3-D结构均由蛋白质数据(Protein Data Bank, PDB) 中获得的相似结构模板和晶体结构组成,所有预测的BvHAK蛋白的C得分在–1.92−1.13之间,表明预测模型的准确性较高。3-D结构进一步验证了对BvHAK基因家族蛋白质的保守基序预测的准确性。
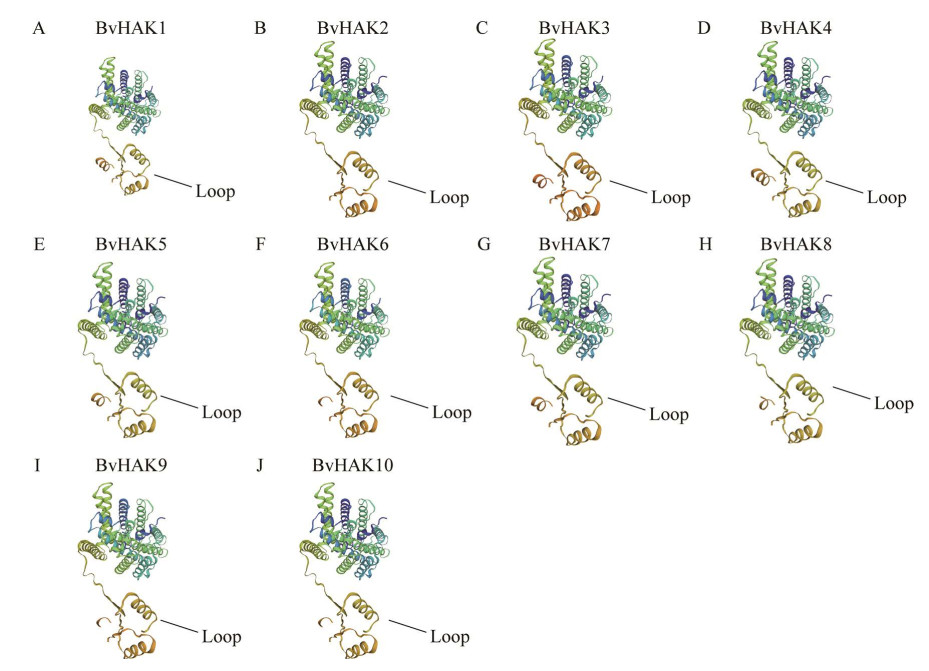
|
| 图 5 甜菜BvHAK家族的蛋白质三维结构 Fig. 5 Three-dimensional structure of BvHAK family proteins. |
| |
采用在线软件STRING对甜菜BvHAK基因家族成员蛋白互作网络进行预测(图 6)。结果表明,8个BvHAK (BvHAK2–BvHAK9) 均与AKT1 (XP_01681784.1) 存在互作关系。BvHAK1、-2、-6、-8和-9与TPK3 (XP_01666502.1) 存在互作。BvHAK1和BvHAK3还与CIPK23 (XP_010687109.1) 存在互作;与BvHAK4存在互作的蛋白有AHA1 (XP_010678592.1)、PME12 (XP_010687598.1)、PME22 (XP_010674080.1)、PME36 (XP_010692199.1)、AHA7 (XP_010692042.1)、BvHKT1; 1 (XP_010688439.1)、BvHKT1; 2 (XP_ 010690257.1) 和BvHKT1; 3 (XP_010690256.1) (图 6)。这些结果为深入研究BvHAK与其他蛋白质的互作及其协同调控网络提供依据。
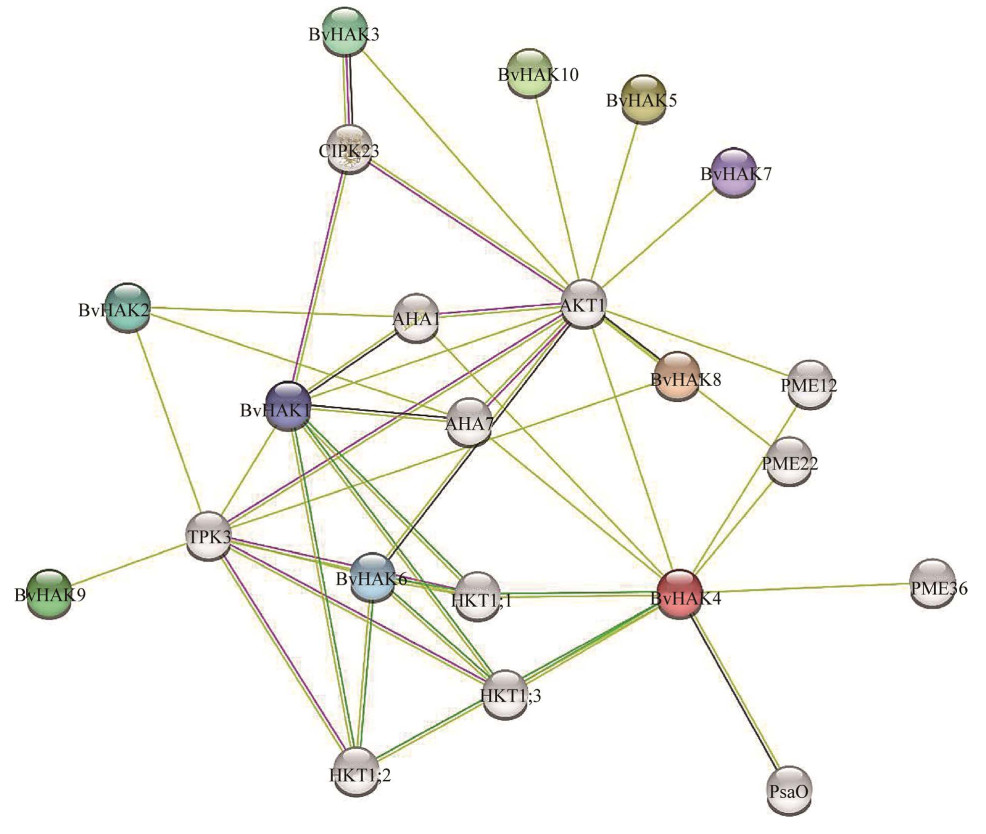
|
| 图 6 甜菜BvHAK家族蛋白互作网络预测 Fig. 6 Predication of protein-protein interaction network of BvHAK family. |
| |
为更全面地了解调控基因表达的诱导因素和基因功能,对甜菜BvHAK基因家族成员翻译起始位点上游1 500 bp启动子顺式作用元件进行了分析。结果表明,BvHAK基因家族成员含有与激素响应元件(如脱落酸、赤霉素、乙烯、生长素),胁迫反应相关元件(如低温、高盐、干旱) 和生长与发育相关响应元件(如玉米醇溶蛋白代谢调节、分生组织和光响应) (表 4)。在激素相关响应元件中,只在BvHAK9中发现了1个GARE-motif;大多数BvHAK (除BvHAK2、-7和-9) 含有1−3个ERE;ABRE在BvHAK3、-4、-6、-9和-10基因的启动子中存在。在胁迫反应相关作用元件中,9个成员(除BvHAK4) 中含有1−3个ARE元件;MYB在9个成员(除BvHAK3) 的启动子中存在。另外,BvHAK1、-3、-5、-6、-9和-10启动子中含有STRE。在生长与发育相关响应元件中,光响应元件无论从类型或数量,在BvHAK基因上游调控区中都是最多的,如box4在8个BvHAK成员(除BvHAK4和BvHAK5) 启动子中均有发现。如表 4所示,BvHAK基因9个成员(除BvHAK2) 含有激素相关响应元件;而胁迫反应和生长与发育相关响应元件存在于所有成员中。特别值得一提的是,BvHAK1和-5含有所有胁迫反应相关响应元件。
| Functional class | Element name | Element function | BvHAK1 | BvHAK2 | BvHAK3 | BvHAK4 | BvHAK5 | BvHAK6 | BvHAK7 | BvHAK8 | BvHAK9 | BvHAK10 |
| Hormone | GARE-motif | Gibberellin-responsive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ABRE | Abscisic acid responsiveness | 0 | 0 | 2 | 3 | 0 | 3 | 0 | 0 | 3 | 1 | |
| TCA-element | Salicylic acid responsiveness | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | |
| TGA-element | Auxin-responsive element | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| ERE | Ethylene-responsive element | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 1 | 0 | 3 | |
| Stress | LTR | Low-temperature responsiveness | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| MYB | Drought related element | 5 | 1 | 0 | 2 | 1 | 3 | 2 | 2 | 3 | 1 | |
| MBS | Drought-inducibility | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |
| ARE | Anaerobic induction | 1 | 1 | 1 | 0 | 3 | 1 | 1 | 1 | 1 | 2 | |
| STRE | Stress response element | 3 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | |
| W-box | Salt-responsive element | 2 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 1 | 2 | |
| Development | AE-box | A module for light response | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Box4 | Light responsiveness | 3 | 5 | 3 | 0 | 0 | 2 | 4 | 4 | 2 | 1 | |
| GCN4_motif | Endosperm expression | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| CAT-box | Meristem expression | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | |
| O2-site | Zein metabolism regulation | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| WRE3 | Wnt responsive DNA element | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 |
为了解甜菜BvHAK基因家族在不同浓度盐处理下表达量变化情况,采用qRT-PCR对其基因表达水平进行相对定量检测。结果显示,与对照(0 mmol/L NaCl) 相比,50和100 mmol/L NaCl显著增加了甜菜叶和根中BvHAK1、BvHAK2、BvHAK4、BvHAK7和BvHAK9表达量(图 7)。在50和100 mmol/L NaCl处理下,叶中的BvHAK2表达量比根中的表达量分别高87.1%和182.9%,BvHAK4在叶中的表达水平比根中的分别高147.5%和53.9%;然而,BvHAK9在叶中的表达量则显著低于根,分别低26.9%和27.7%。在50 mmol/L NaCl处理下,BvHAK7在根中的表达量高于叶;而在100 mmol/L处理下,BvHAK7在叶中的表达量显著高于根。与对照相比,高盐(150 mmol/L NaCl) 处理显著降低了叶和根中BvHAK1和BvHAK7的表达量。另外,高盐处理显著降低了BvHAK6、BvHAK8和BvHAK9在叶中的表达量,而对根中的表达量影响差异不显著。这些结果表明,50和100 mmol/L NaCl不同程度地诱导甜菜叶和根中BvHAK基因家族成员的表达;高盐则下调了BvHAK在叶中的表达水平,而对根中的表达量没有影响。
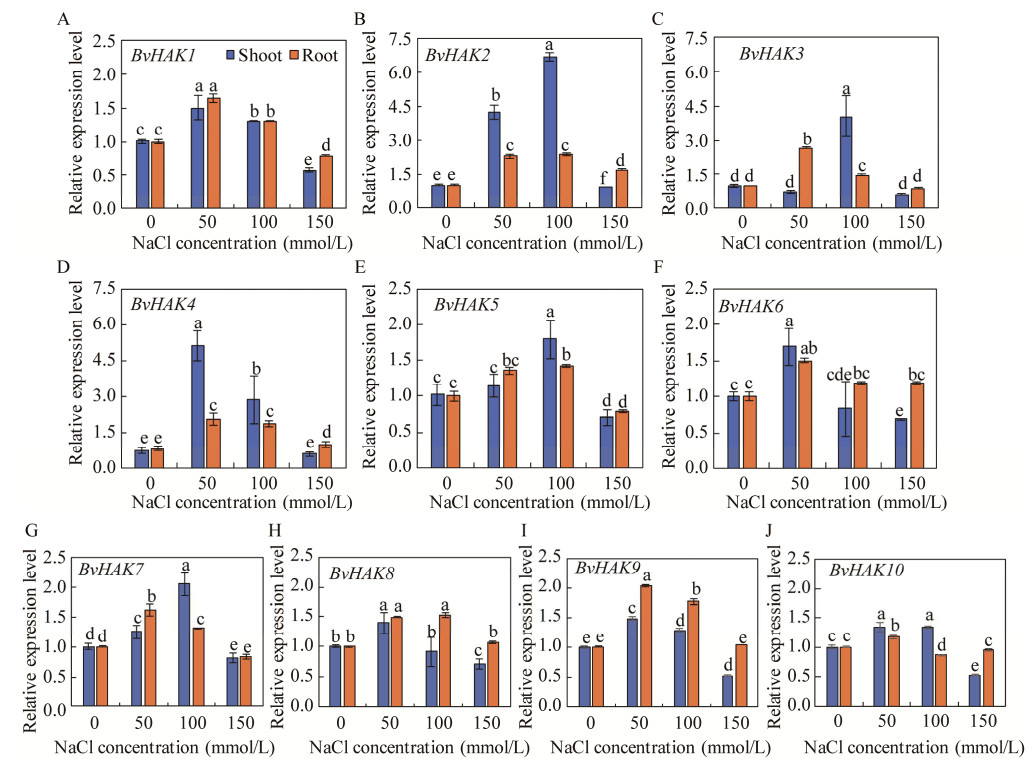
|
| 图 7 甜菜BvHAK基因家族成员在不同浓度NaCl处理下的相对表达水平 Fig. 7 Relative expression levels of BvHAK gene family members under various concentrations of NaCl. (A) Relative expression levels of BvHAK1 in shoots and roots under different concentrations of NaCl. (B): Relative expression levels of BvHAK2 in shoots and roots under different concentrations of NaCl. (C): Relative expression levels of BvHAK3 in shoots and roots under different concentrations of NaCl. (D): Relative expression levels of BvHAK4 in shoots and roots under different concentrations of NaCl; (E): Relative expression levels of BvHAK5 in shoots and roots under different concentrations of NaCl. (F): Relative expression levels of BvHAK6 in shoots and roots under different concentrations of NaCl. (G): Relative expression levels of BvHAK7 in shoots and roots under different concentrations of NaCl. (H): Relative expression levels of BvHAK8 in shoots and roots under different concentrations of NaCl. (I): Relative expression levels of BvHAK9 in shoots and roots under different concentrations of NaCl. (J): Relative expression levels of BvHAK10 in shoots and roots under different concentrations of NaCl. Different letters mean significant difference (P < 0.05). A:不同浓度NaCl处理下BvHAK1在叶和根中的相对表达水平;B:不同浓度NaCl处理下BvHAK2在叶和根中的相对表达水平;C:不同浓度NaCl处理下BvHAK3在叶和根中的相对表达水平;D:不同浓度NaCl处理下BvHAK4在叶和根中的相对表达水平;E:不同浓度NaCl处理下BvHAK5在叶和根中的相对表达水平;F:不同浓度NaCl处理下BvHAK6在叶和根中的相对表达水平;G:不同浓度NaCl处理下BvHAK7在叶和根中的相对表达水平;H:不同浓度NaCl处理下BvHAK8在叶和根中的相对表达水平;I:不同浓度NaCl处理下BvHAK9在叶和根中的相对表达水平;J:不同浓度NaCl处理下BvHAK10在叶和根中的相对表达水平。不同字母表示差异显著(P < 0.05) |
| |
在本研究中,甜菜基因组数据库中共鉴定出10个BvHAK基因(表 2),与其他物种的HAK基因家族成员数量不尽相同,如拟南芥中有13个[28],玉米27个、马铃薯15个、水稻27个[45],桃(Prunus persica) 15个,葡萄18个[46]、红皮柳(Salix purpurea) 22个[47]、茶树(Camellia sinensis) 21个[48]。可能是由于在进化过程中HAK家族不同亚科的特定基因复制和损失,造成了植物中HAK家族基因数量上的差异。
根据亚细胞定位预测,6个BvHAK基因家族成员定位在液泡膜(BvHAK1、-4、-6、-7、-9和-10),而4个成员定位在质膜(BvHAK2、-3、-5和-8)。已有研究表明,绝大多数植物HAK基因家族成员定位于质膜上[49]。在水稻中,OsHAK10定位于液泡膜上,也有部分HAK家族成员如小立碗藓(Physcomitrella patens) PpHAK2定位于内膜,PpHAK3定位于高尔基体[50]。内含子与外显子的数目和结构也是基因家族进化中的一项重要指标。在本研究中,3个BvHAK基因家族成员(BvHAK3、-7和-10) 含有8个外显子,5个成员(BvHAK1、-4、-5、-6和-8) 含有9个外显子,其余BvHAK家族成员含有10个外显子(图 1A)。红皮柳SpuHAK13含有6个外显子,梨(Pyrus bretschneideri) PbrHAK11中有7个外显子,PbrHAK7中有6个外显子[51]。高等植物中的大部分KT/HAK/KUP基因包含8−10个外显子(少于绿藻中的外显子)[52]。对于HAK蛋白家族成员,K+转运蛋白结构域存在于所有物种中,具有高度的相似性。这些结果表明,高等植物HAK基因家族序列相对保守。
顺式作用元件参与基因活性调控,是转录调控的关键分子开关[53]。在植物中,生长素ABA、乙烯等激素在植物的生长发育与逆境胁迫响应过程中起着重要作用[54]。在BvHAK基因家族成员中,鉴定出39个顺式作用调控元件,其中激素响应元件7个,胁迫响应元件8个,生长发育元件24个(表 4)。在互作网络预测研究中,除BvHAK2与BvHAK9外,其余BvHAK蛋白均与向内整流通道AKT1互作[55]。另外,BvHAK家族成员与K+通道TPK3互作[56]。在本研究中,BvHAK3还与CIPK23相互作用(图 6)。在辣椒(Capsicum annuum) 和番茄中,HAK5及同源蛋白可被CBL1/CIPK23复合物激活[57-58]。在甘蔗(Saccharum officinarum) 中,SsHAK1可能被CBL1-CIPK23复合物或RUPO磷酸化和激活[59]。这些研究结果为进一步揭示HAK调控机制奠定基础。
3.2 甜菜BvHAK在盐处理下的表达模式分析在本研究中,盐处理显著诱导了BvHAK4在叶和根中的表达水平。研究表明,水稻OsHAK2介导Na+转运和低亲和性K+转运,大麦HvHAK2为低亲和性K+转运且对Na+敏感。棉花(Gossypium hirsutum) GhHKT2[60]、甘蔗SsHAK2、葡萄VvKUP2中也均发现有类似功能。拟南芥AtHAK5主要在根中表达,介导K+的吸收[51]。水稻OsHAK5则为Na+敏感型的高亲和性K+转运蛋白[12]。在水稻根和叶中均检测到OsHAK5表达,其通过增强根中K+的吸收和K+从根至地上部的转运,从而提高K+/Na+比,保持离子稳态,增强水稻的耐盐性[61]。芦苇(Phragmites australis) PhaHAK5具有Na+渗透性,也可提高植物的耐盐性[62]。在盐胁迫下,BvHAK3在根和叶中均有表达,但叶中的表达量显著高于根中。BvHAK3在低盐胁迫处理后,根中的表达量达到峰值;但在100 mmol/L NaCl处理后,叶中的表达量达到最高。另外,小立碗藓PpHAK13主要介导Na+转运[63]。系统发育分析表明,BvHAK6与拟南芥AtHAK12进化关系较近(图 2),其功能可能与AtKHAK12功能相似,通过光合系统参与甜菜对盐胁迫的响应。玉米ZmHAK4在根的维管束中优先表达,编码一种新的质膜Na+选择性转运蛋白,该转运蛋白可能通过从木质部汁液中回收Na+,从而介导地上部Na+外排[64]。在水稻和小麦中,与玉米ZmHAK4的同源基因,具有相同的表达模式和离子转运特性,表明ZmHAK4及其同源基因具有相似的耐盐机制[35]。在本研究中,BvHAK4在50 mmol/L NaCl处理下地上部表达量最高(图 7D),推测其与ZmHAK4具有相同的耐盐机理。另外,BvHAK5在50 mmol/L NaCl处理后叶表现出较高的表达水平(图 7E),这与拟南芥中AtKUP7的表达模式一致。由此说明,BvHAK5对甜菜根系吸收K+至关重要,且可能参与K+向木质部汁液的转运[65]。
4 结论在本研究中,共鉴定10个甜菜BvHAK基因家族成员,其含有8−10个外显子、7−9个内含子不等;平均氨基酸个数为778.3,平均分子量为88.31 kDa,等电点为5.38−9.41,跨膜区为11−14个不等。系统进化分析发现,高等植物HAK可分为5个簇,分别为Ⅰ、Ⅱ、Ⅲ、Ⅴ和Ⅳ簇,其中Ⅱ簇成员可进一步分为Ⅱa、Ⅱb和Ⅱc等3个亚簇;BvHAK家族成员则分布在前4簇,Ⅰ簇和Ⅲ簇有1个成员,Ⅱ簇有6个成员,Ⅴ簇有2个成员。进一步对BvHAK基因在盐处理下甜菜不同组织中的表达模式分析发现,50和100 mmol/L NaCl不同程度地诱导甜菜叶和根中BvHAK基因家族成员的表达;高盐(150 mmol/L) 则下调其在叶中的表达水平,而对根中的表达量没有影响。
| [1] |
Jorge TF, Tohge T, Wendenburg R, et al. Salt-stress secondary metabolite signatures involved in the ability of Casuarina glauca to mitigate oxidative stress. Environ Exp Bot, 2019, 166: 103808. DOI:10.1016/j.envexpbot.2019.103808
|
| [2] |
Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol, 2008, 59: 651-681. DOI:10.1146/annurev.arplant.59.032607.092911
|
| [3] |
杨真, 王宝山. 中国盐渍土资源现状及改良利用对策. 山东农业科学, 2015, 47(4): 125-130. Yang Z, Wang BS. Present status of saline soil resources and countermeasures for improvement and utilization in China. Shandong Agric Sci, 2015, 47(4): 125-130 (in Chinese). DOI:10.14083/j.issn.1001-4942.2015.04.032 |
| [4] |
Etesami H, Glick BR. Halotolerant plant growth-promoting bacteria: prospects for alleviating salinity stress in plants. Environ Exp Bot, 2020, 178: 104124. DOI:10.1016/j.envexpbot.2020.104124
|
| [5] |
Assaha DVM, Ueda A, Saneoka H, et al. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol, 2017, 8: 509. DOI:10.3389/fphys.2017.00509
|
| [6] |
Rajappa S, Krishnamurthy P, Kumar PP. Regulation of AtKUP2 expression by bHLH and WRKY transcription factors helps to confer increased salt tolerance to Arabidopsis thaliana plants. Front Plant Sci, 2020, 11: 1311. DOI:10.3389/fpls.2020.01311
|
| [7] |
Alqahtani M, Lightfoot DJ, Lemtiri-Chlieh F, et al. The role of PQL genes in response to salinity tolerance in Arabidopsis and barley. Plant Direct, 2021, 5(2): e00301.
|
| [8] |
Gupta A, Shaw BP, Roychoudhury A. NHX1, HKT, and monovalent cation transporters regulate K+ and Na+ transport during abiotic stress. Transporters and Plant Osmotic Stress. Amsterdam: Elsevier, 2021, 1-27.
|
| [9] |
Zhang HW, Xiao W, Yu WW, et al. Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low K+ or salt stress. Plant Cell Rep, 2018, 37(11): 1533-1546. DOI:10.1007/s00299-018-2325-2
|
| [10] |
Li WH, Xu GH, Alli A, et al. Plant HAK/KUP/KT K+ transporters: function and regulation. Semin Cell Dev Biol, 2018, 74: 133-141. DOI:10.1016/j.semcdb.2017.07.009
|
| [11] |
Gobert A, Park G, Amtmann A, et al. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J Exp Bot, 2006, 57(4): 791-800. DOI:10.1093/jxb/erj064
|
| [12] |
Horie T, Sugawara M, Okada T, et al. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioeng, 2011, 111(3): 346-356. DOI:10.1016/j.jbiosc.2010.10.014
|
| [13] |
晁毛妮, 温青玉, 张晋玉, 等. 大豆KUP/HAK/KT钾转运体基因家族的鉴定与表达分析. 西北植物学报, 2017, 37(2): 239-249. Chao MN, Wen QY, Zhang JY, et al. Identification and expression analysis of KUP/HAK/KT potassium transporter gene family in soybean [Glycine max (L.) Merr]. Acta Bot Boreali Occidentalia Sin, 2017, 37(2): 239-249 (in Chinese). |
| [14] |
马丽娟, 马钰, 何红红, 等. 葡萄HAK基因家族的鉴定与表达分析. 西北农业学报, 2019, 28(6): 922-934. Ma LJ, Ma Y, He HH, et al. Identification and expression analysis of grape HAK gene family. Acta Agric Boreali Occidentalis Sin, 2019, 28(6): 922-934 (in Chinese). |
| [15] |
肖小虎, 林显祖, 龙翔宇, 等. 橡胶树KT/HAK/KUP基因家族成员的鉴定与表达分析. 热带作物学报, 2022, 43(1): 1-8. Xiao XH, Lin XZ, Long XY, et al. Identifcation and expression of KT/HAK/KUP genes in Hevea brasliensis. Chin J Trop Crops, 2022, 43(1): 1-8 (in Chinese). |
| [16] |
许赛赛, 张博, 仲阳, 等. 马铃薯HAK/KUP/KT基因家族鉴定与表达分析. 分子植物育种, 2021, 19(12): 3878-3886. Xu SS, Zhang B, Zhong Y, et al. Identification and expression analysis of HAK/KUP/KT gene family in potato. Mol Plant Breed, 2021, 19(12): 3878-3886 (in Chinese). |
| [17] |
田娜, 王斌, 伍俊为, 等. 香蕉高亲和性钾转运蛋白(HAK) 家族全基因组鉴定及表达. 应用与环境生物学报, 2021. Tian N, Wang B, Wu JW, et al. Genome-wide identification and expression of the banana high-affinity potassium transporter (HAK) family. Chin J Appl Environ Biol, 2021 (in Chinese). DOI:10.19675/j.cnki.1006-687x.2021.04010 |
| [18] |
朱乐, 赵鑫泽, 蒋立希. 甘蓝型油菜钾离子转运载体HAK/KUP/KT家族的全基因组鉴定与分析. 浙江大学学报(农业与生命科学版), 2021, 47(3): 303-313. Zhu L, Zhao XZ, Jiang LX. Genome-wide identification and analysis of potassium ion transporter HAK/KUP/KT family in rapeseed (Brassica napus L.). J Zhejiang Univ (Agric Life Sci), 2021, 47(3): 303-313 (in Chinese). |
| [19] |
吴胜男, 杨媛, 李英壮, 等. 小麦KUP/HAK/KT基因家族的全基因组鉴定、系统进化和表达模式分析. 西北农业学报, 2021, 30(3): 351-364. Wu SN, Yang Y, Li YZ, et al. Genome-wide identification, phylogeny and expression analysis of KUP/HAK/KT transcription factors in wheat. Acta Agric Boreali Occidentalis Sin, 2021, 30(3): 351-364 (in Chinese). |
| [20] |
Leiva-Eriksson N, Pin PA, Kraft T, et al. Differential expression patterns of non-symbiotic hemoglobins in sugar beet (Beta vulgaris ssp. vulgaris). Plant Cell Physiol, 2014, 55(4): 834-844. DOI:10.1093/pcp/pcu027
|
| [21] |
Wu GQ, Feng RJ, Liang N, et al. Sodium chloride stimulates growth and alleviates sorbitol-induced osmotic stress in sugar beet seedlings. Plant Growth Regul, 2015, 75(1): 307-316. DOI:10.1007/s10725-014-9954-4
|
| [22] |
Dohm JC, Minoche AE, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature, 2014, 505(7484): 546-549. DOI:10.1038/nature12817
|
| [23] |
Wu GQ, Wang JL, Li SJ. Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes, 2019, 10(5): 401. DOI:10.3390/genes10050401
|
| [24] |
Wu GQ, Li ZQ, Cao H, et al. Genome-wide identification and expression analysis of the WRKY genes in sugar beet (Beta vulgaris L.) under alkaline stress. PeerJ, 2019, 7: e7817. DOI:10.7717/peerj.7817
|
| [25] |
Wu GQ, Liu ZX, Xie LL, et al. Genome-wide identification and expression analysis of the BvSnRK2 genes family in sugar beet (Beta vulgaris L.) under salt conditions. J Plant Growth Regul, 2021, 40(2): 519-532. DOI:10.1007/s00344-020-10119-y
|
| [26] |
Wu GQ, Xie LL, Wang JL, et al. Genome-wide identification of CIPK genes in sugar beet (Beta vulgaris) and their expression under NaCl stress. J Plant Growth Regul, 2022, DOI: org/10.1007/s00344-021-10545-6.
|
| [27] |
Genies L, Orjollet D, Carasco L, et al. Uptake and translocation of cesium by Arabidopsis thaliana in hydroponics conditions: links between kinetics and molecular mechanisms. Environ Exp Bot, 2017, 138: 164-172. DOI:10.1016/j.envexpbot.2017.03.013
|
| [28] |
Quintero FJ, Blatt MR. A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett, 1997, 415(2): 206-211. DOI:10.1016/S0014-5793(97)01125-3
|
| [29] |
Del Río ÁR, Minoche AE, Zwickl NF, et al. Genomes of the wild beets Beta patula and Beta vulgaris ssp. Maritima. Plant J, 2019, 99(6): 1242-1253. DOI:10.1111/tpj.14413
|
| [30] |
Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook.. Totowa, NJ: Humana Press, 2005: 571-607.
|
| [31] |
Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One, 2010, 5(6): e11335. DOI:10.1371/journal.pone.0011335
|
| [32] |
Azeem F, Zameer R, Rehman Rashid MA, et al. Genome-wide analysis of potassium transport genes in Gossypium raimondii suggest a role of GrHAK/KUP/KT8, GrAKT2.1 and GrAKT1. 1 in response to abiotic stress. Plant Physiol Biochem, 2022, 170: 110-122. DOI:10.1016/j.plaphy.2021.11.038
|
| [33] |
Jin R, Jiang W, Yan MX, et al. Genome-wide characterization and expression analysis of HAK K+ transport family in Ipomoea. 3 Biotech, 2021, 11(1): 3. DOI:10.1007/s13205-020-02552-3
|
| [34] |
Zhang M, Liang XY, Wang LM, et al. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat Plants, 2019, 5(12): 1297-1308. DOI:10.1038/s41477-019-0565-y
|
| [35] |
Cai KF, Zeng FR, Wang JM, et al. Identification and characterization of HAK/KUP/KT potassium transporter gene family in barley and their expression under abiotic stress. BMC Genomics, 2021, 22(1): 317. DOI:10.1186/s12864-021-07633-y
|
| [36] |
Cheng XY, Liu XD, Mao WW, et al. Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). Int J Mol Sci, 2018, 19(12): 3969. DOI:10.3390/ijms19123969
|
| [37] |
Kumar S, Stecher G, Tamura K. MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016, 33(7): 1870-1874. DOI:10.1093/molbev/msw054
|
| [38] |
Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol, 2007, 24(8): 1586-1591. DOI:10.1093/molbev/msm088
|
| [39] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 2015, 31(8): 1296-1297. DOI:10.1093/bioinformatics/btu817
|
| [40] |
Guan Y, Wang DH, Lv C, et al. Archives of microbiology: screening of pectinase-producing bacteria from Citrus peel and characterization of a recombinant pectate lyase with applied potential. Arch Microbiol, 2020, 202(5): 1005-1013. DOI:10.1007/s00203-020-01807-0
|
| [41] |
Costa CS, Bravo JP, Ribeiro CL, et al. Vascular expression driven by the promoter of a gene encoding a high-affinity potassium transporter HAK5 from Eucalyptus grandis. Plant Cell Tissue Organ Cult, 2017, 131(2): 213-222. DOI:10.1007/s11240-017-1276-6
|
| [42] |
Oliferuk S, Simontacchi M, Rubio F, et al. Exposure to a natural nitric oxide donor negatively affects the potential influx of rubidium in potassium-starved Arabidopsis plants. Plant Physiol Biochem, 2020, 150: 204-208. DOI:10.1016/j.plaphy.2020.02.043
|
| [43] |
Dai FB, Li AJ, Rao SP, et al. Potassium transporter LrKUP8 is essential for K+ preservation in Lycium ruthenicum, a salt-resistant desert shrub. Genes, 2019, 10(8): 600. DOI:10.3390/genes10080600
|
| [44] |
Sun Q, Lu HR, Zhang Q, et al. Transcriptome sequencing of wild soybean revealed gene expression dynamics under low nitrogen stress. J Appl Genet, 2021, 62(3): 389-404. DOI:10.1007/s13353-021-00628-1
|
| [45] |
Gupta M, Qiu XH, Wang L, et al. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genom, 2008, 280(5): 437-452. DOI:10.1007/s00438-008-0377-7
|
| [46] |
Davies C, Shin R, Liu WH, et al. Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J Exp Bot, 2006, 57(12): 3209-3216. DOI:10.1093/jxb/erl091
|
| [47] |
Liang MX, Gao YC, Mao TT, et al. Characterization and expression of KT/HAK/KUP transporter family genes in willow under potassium deficiency, drought, and salt stresses. Biomed Res Int, 2020, 2020: 2690760.
|
| [48] |
Yang TY, Lu X, Wang Y, et al. HAK/KUP/KT family potassium transporter genes are involved in potassium deficiency and stress responses in tea plants (Camellia sinensis L.): expression and functional analysis. BMC Genomics, 2020, 21(1): 556. DOI:10.1186/s12864-020-06948-6
|
| [49] |
Li Y, Peng LR, Xie CY, et al. Genome-wide identification, characterization, and expression analyses of the HAK/KUP/KT potassium transporter gene family reveals their involvement in K+ deficient and abiotic stress responses in pear rootstock seedlings. Plant Growth Regul, 2018, 85(2): 187-198. DOI:10.1007/s10725-018-0382-8
|
| [50] |
Haro R, Fraile-Escanciano A, González-Melendi P, et al. The potassium transporters HAK2 and HAK3 localize to endomembranes in Physcomitrella patens.HAK2 is required in some stress conditions. Plant Cell Physiol, 2013, 54(9): 1441-1454. DOI:10.1093/pcp/pct097
|
| [51] |
Gierth M, Mäser P, Schroeder JI. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol, 2005, 137(3): 1105-1114. DOI:10.1104/pp.104.057216
|
| [52] |
He CY, Cui K, Duan AG, et al. Genome-wide and molecular evolution analysis of the Poplar KT/HAK/KUP potassium transporter gene family. Ecol Evol, 2012, 2(8): 1996-2004. DOI:10.1002/ece3.299
|
| [53] |
Ding X, Li JH, Pan Y, et al. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int J Mol Sci, 2018, 19(6): 1583.
|
| [54] |
Zhang YL, Li X. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol, 2019, 50: 29-36.
|
| [55] |
Nieves-Cordones M, Lara A, Ródenas R, et al. Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell Environ, 2019, 42(8): 2357-2371.
|
| [56] |
Adams E, Miyazaki T, Shin R. Contribution of KUPs to potassium and cesium accumulation appears complementary in Arabidopsis. Plant Signal Behav, 2019, 14(1): 1554468.
|
| [57] |
Ragel P, Ródenas R, García-Martín E, et al. The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol, 2015, 169(4): 2863-2873.
|
| [58] |
Martínez-Cordero MA, Martínez V, Rubio F. Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol Biol, 2004, 56(3): 413-421.
|
| [59] |
Feng XM, Wang YJ, Zhang NN, et al. Genome-wide systematic characterization of the HAK/KUP/KT gene family and its expression profile during plant growth and in response to low-K+ stress in Saccharum. BMC Plant Biol, 2020, 20(1): 20.
|
| [60] |
Li Z, Tian X, Zhang M, et al. GhKT2: a novel K+ transporter gene in cotton (Gossypium hirsutum). Front Agr Sci Eng, 2018, 5(2): 226-235.
|
| [61] |
Feng HM, Tang Q, Cai J, et al. Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta, 2019, 250(2): 549-561.
|
| [62] |
Takahashi R, Nishio T, Ichizen N, et al. High-affinity K+ transporter PhaHAK5 is expressed only in salt- sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Rep, 2007, 26(9): 1673-1679.
|
| [63] |
Benito B, Garciadeblas B, Rodriguez-Navarro A. HAK transporters from Physcomitrella patens and Yarrowia lipolytica mediate sodium uptake. Plant Cell Physiol, 2012, 53(6): 1117-1123.
|
| [64] |
Ma NN, Dong LN, Lü W, et al. Transcriptome analysis of maize seedling roots in response to nitrogen-, phosphorus-, and potassium deficiency. Plant Soil, 2020, 447(1/2): 637-658.
|
| [65] |
Šustr M, Doksanská T, Doležalová B, et al. 134Cs uptake and growth at various Cs+ and K+ levels in Arabidopsis AtKUP7 mutants. Plants, 2020, 9(11): 1525.
|
 2022, Vol. 38
2022, Vol. 38