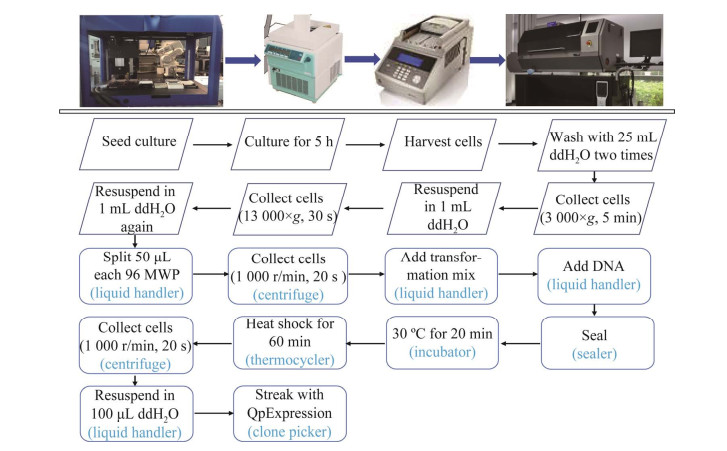
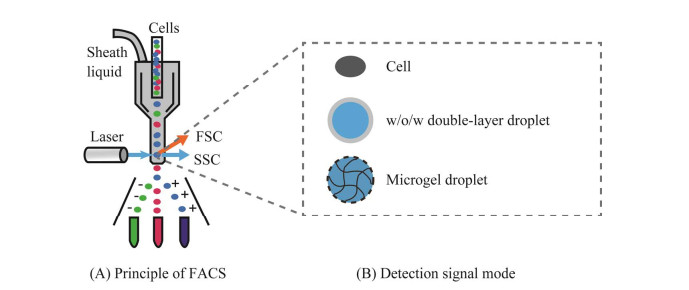
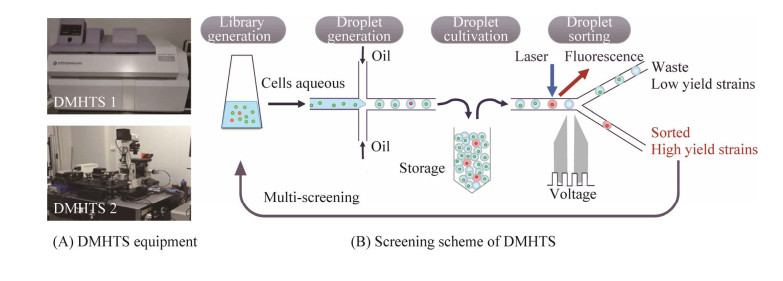
| [1] |
Clomburg JM, Crumbley AM, Gonzalez R. Industrial biomanufacturing: the future of chemical production. Science, 2017, 355(6320): aag0804. DOI:10.1126/science.aag0804
|
|
| [2] |
唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台. 科学通报, 2021, 66(3): 300-309. Tang T, Fu LH, Guo EP, et al. Automation in synthetic biology using biological foundries. Chin Sci Bull, 2021, 66(3): 300-309 (in Chinese).
|
|
| [3] | |
|
| [4] | |
|
| [5] | |
|
| [6] |
Hillson NJ, Rosengarten RD, Keasling JD. J5 DNA assembly design automation software. ACS Synth Biol, 2012, 1(1): 14-21. DOI:10.1021/sb2000116
|
|
| [7] |
Enghiad B, Xue P, Singh N, et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction. Nat Commun, 2022, 13(1): 2697. DOI:10.1038/s41467-022-30355-y
|
|
| [8] |
Wang Y, Liu Y, Li JW, et al. Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum. Biotechnol Bioeng, 2019, 116(11): 3016-3029. DOI:10.1002/bit.27121
|
|
| [9] |
Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res, 2014, 42(W1): W401-W407. DOI:10.1093/nar/gku410
|
|
| [10] |
Cárdenas P, Esherick LY, Chambonnier G, et al. GeneTargeter: automated in silico design for genome editing in the malaria parasite, Plasmodium falciparum. The CRISPR Journal, 2022, 5(1): 155-164. DOI:10.1089/crispr.2021.0069
|
|
| [11] |
Yang Y, Mao YF, Liu Y, et al. GEDpm-cg: genome editing automated design platform for point mutation construction in Corynebacterium glutamicum. Front Bioeng Biotechnol, 2021, 9: 768289. DOI:10.3389/fbioe.2021.768289
|
|
| [12] |
Yang Y, Mao YF, Wang RY, et al. AutoESD: a web tool for automatic editing sequence design for genetic manipulation of microorganisms. Nucleic Acids Res, 2022, 50(W1): W75-W82. DOI:10.1093/nar/gkac417
|
|
| [13] |
Storch M, Haines MC, Baldwin GS. DNA-BOT: a low-cost, automated DNA assembly platform for synthetic biology. Synthetic biology (Oxford, England), 2020, 5(1): ysaa010. DOI:10.1093/synbio/ysaa010
|
|
| [14] |
Casini A, Chang FY, Eluere R, et al. A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J Am Chem Soc, 2018, 140(12): 4302-4316. DOI:10.1021/jacs.7b13292
|
|
| [15] |
Si T, Chao R, Min YH, et al. Automated multiplex genome-scale engineering in yeast. Nat Commun, 2017, 8(1): 15187. DOI:10.1038/ncomms15187
|
|
| [16] |
Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng, 2018, 50: 122-141. DOI:10.1016/j.ymben.2018.07.008
|
|
| [17] |
Wang Y, Liu Y, Liu J, et al. MACBETH: multiplex automated Corynebacterium glutamicum base editing method. Metab Eng, 2018, 47: 200-210. DOI:10.1016/j.ymben.2018.02.016
|
|
| [18] |
Park SA, Bhatia SK, Park HA, et al. Bacillus subtilis as a robust host for biochemical production utilizing biomass. Crit Rev Biotechnol, 2021, 41(6): 827-848. DOI:10.1080/07388551.2021.1888069
|
|
| [19] |
Yu SL, Price MA, Wang Y, et al. CRISPR-dCas9 mediated cytosine deaminase base editing in Bacillus subtilis. ACS Synth Biol, 2020, 9(7): 1781-1789. DOI:10.1021/acssynbio.0c00151
|
|
| [20] |
Lian JZ, Mishra S, Zhao HM. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab Eng, 2018, 50: 85-108. DOI:10.1016/j.ymben.2018.04.011
|
|
| [21] |
Lan XT, Yuan W, Wang M, et al. Efficient biosynthesis of antitumor ganoderic acid HLDOA using a dual tunable system for optimizing the expression of CYP5150L8 and a Ganoderma P450 reductase. Biotechnol Bioeng, 2019, 116(12): 3301-3311. DOI:10.1002/bit.27154
|
|
| [22] |
涂然, 李世新, 李昊霓, 等. 液滴微流控技术在微生物工程菌株选育中的应用进展. 合成生物学, 2022, 3: 1-21.
Tu R, Li SX, Li HN, et al. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains. Syn Bio J, 2022, 3: 1-21 (in Chinese).
|
|
| [23] |
Leavell MD, Singh AH, Kaufmann-Malaga BB. High-throughput screening for improved microbial cell factories, perspective and promise. Curr Opin Biotechnol, 2020, 62: 22-28. DOI:10.1016/j.copbio.2019.07.002
|
|
| [24] |
Liu Y, Liu Y, Wang M. Design, optimization and application of small molecule biosensor in metabolic engineering. Front Microbiol, 2017, 8: 2012. DOI:10.3389/fmicb.2017.02012
|
|
| [25] |
Schallmey M, Frunzke J, Eggeling L, et al. Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors. Curr Opin Biotechnol, 2014, 26: 148-154. DOI:10.1016/j.copbio.2014.01.005
|
|
| [26] |
Wang Y, Li QG, Zheng P, et al. Evolving the L-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method. J Ind Microbiol Biotechnol, 2016, 43(9): 1227-1235. DOI:10.1007/s10295-016-1803-1
|
|
| [27] |
Liu YN, Li QG, Zheng P, et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb Cell Fact, 2015, 14: 121. DOI:10.1186/s12934-015-0311-8
|
|
| [28] |
Li LP, Tu R, Song GT, et al. Development of a synthetic 3-dehydroshikimate biosensor in Escherichia coli for metabolite monitoring and genetic screening. ACS Synth Biol, 2019, 8(2): 297-306. DOI:10.1021/acssynbio.8b00317
|
|
| [29] |
Liu YF, Zhuang YY, Ding DQ, et al. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synth Biol, 2017, 6(5): 837-848. DOI:10.1021/acssynbio.6b00328
|
|
| [30] |
Han GQ, Xu N, Sun XP, et al. Improvement of L-valine production by atmospheric and room temperature plasma mutagenesis and high-throughput screening in Corynebacterium glutamicum. ACS Omega, 2020, 5(10): 4751-4758. DOI:10.1021/acsomega.9b02747
|
|
| [31] |
Ding D, Li J, Bai D, et al. Biosensor-based monitoring of the central metabolic pathway metabolites. Biosens Bioelectron, 2020, 167: 112456. DOI:10.1016/j.bios.2020.112456
|
|
| [32] |
Liu YF, Yuan HL, Ding DQ, et al. Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction. ACS Synth Biol, 2021, 10(6): 1373-1383. DOI:10.1021/acssynbio.0c00647
|
|
| [33] |
Sun X, Li QG, Wang Y, et al. Isoleucyl-tRNA synthetase mutant based whole-cell biosensor for high-throughput selection of isoleucine overproducers. Biosens Bioelectron, 2021, 172: 112783. DOI:10.1016/j.bios.2020.112783
|
|
| [34] |
Ma FQ, Guo TJ, Zhang YF, et al. An ultrahigh-throughput screening platform based on flow cytometric droplet sorting for mining novel enzymes from metagenomic libraries. Environ Microbiol, 2021, 23(2): 996-1008. DOI:10.1111/1462-2920.15257
|
|
| [35] |
Korfer G, Pitzler C, Vojcic L, et al. In vitro flow cytometry-based screening platform for cellulase engineering. Sci Rep, 2016, 6: 26128. DOI:10.1038/srep26128
|
|
| [36] |
Zhu XD, Shi X, Wang SW, et al. High-throughput screening of high lactic acid-producing Bacillus coagulans by droplet microfluidic based flow cytometry with fluorescence activated cell sorting. RSC Adv, 2019, 9(8): 4507-4513. DOI:10.1039/C8RA09684H
|
|
| [37] |
Wagner JM, Liu L, Yuan SF, et al. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes: riboflavin overproduction in Yarrowia lipolytica. Metab Eng, 2018, 47: 346-356. DOI:10.1016/j.ymben.2018.04.015
|
|
| [38] |
Yanakieva D, Elter A, Bratsch J, et al. FACS-based functional protein screening via microfluidic co-encapsulation of yeast secretor and mammalian reporter cells. Sci Rep, 2020, 10(1): 10182. DOI:10.1038/s41598-020-66927-5
|
|
| [39] |
Hernandez-Valdes JA, Aan De Stegge M, Hermans J, et al. Enhancement of amino acid production and secretion by Lactococcus lactis using a droplet-based biosensing and selection system. Metab Eng Commun, 2020, 11: e00133. DOI:10.1016/j.mec.2020.e00133
|
|
| [40] |
Li M, Van Zee M, Riche CT, et al. A gelatin microdroplet platform for high-throughput sorting of hyperproducing single-cell-derived microalgal clones. Small, 2018, 14(44): e1803315. DOI:10.1002/smll.201803315
|
|
| [41] |
Delgado-Ramos L, Marcos AT, Ramos-Guelfo MS, et al. Flow cytometry of microencapsulated colonies for genetics analysis of filamentous fungi. G3 (Bethesda), 2014, 4(11): 2271-2278. DOI:10.1534/g3.114.014357
|
|
| [42] |
Wang GK, Jia WD, Chen N, et al. A GFP-fusion coupling FACS platform for advancing the metabolic engineering of filamentous fungi. Biotechnol Biofuels, 2018, 11: 232. DOI:10.1186/s13068-018-1223-8
|
|
| [43] |
Yang YJ, Liu Y, Liu DD, et al. Development of a flow cytometry-based plating-free system for strain engineering in industrial fungi. Appl Microbiol Biotechnol, 2022, 106(2): 713-727. DOI:10.1007/s00253-021-11733-w
|
|
| [44] |
Bai CX, Zhang Y, Zhao XJ, et al. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. PNAS, 2015, 112(39): 12181-12186. DOI:10.1073/pnas.1511027112
|
|
| [45] |
Tu R, Zhang Y, Hua EB, et al. Droplet-based microfluidic platform for high-throughput screening of Streptomyces. Commun Biol, 2021, 4(1): 647. DOI:10.1038/s42003-021-02186-y
|
|
| [46] |
Yang JH, Tu R, Yuan HL, et al. Recent advances in droplet microfluidics for enzyme and cell factory engineering. Crit Rev Biotechnol, 2021, 41(7): 1023-1045. DOI:10.1080/07388551.2021.1898326
|
|
| [47] |
Bowman EK, Alper HS. Microdroplet-assisted screening of biomolecule production for metabolic engineering applications. Trends Biotechnol, 2020, 38(7): 701-714. DOI:10.1016/j.tibtech.2019.11.002
|
|
| [48] |
Josephides D, Davoli S, Whitley W, et al. Cyto-Mine: an integrated, picodroplet system for high-throughput single-cell analysis, sorting, dispensing, and monoclonality assurance. SLAS Technol, 2020, 25(2): 177-189. DOI:10.1177/2472630319892571
|
|
| [49] |
He RL, Ding RH, Heyman JA, et al. Ultra-high-throughput picoliter-droplet microfluidics screening of the industrial cellulase-producing filamentous fungus Trichoderma reesei. J Ind Microbiol Biotechnol, 2019, 46(11): 1603-1610. DOI:10.1007/s10295-019-02221-2
|
|
| [50] |
Yun KY, Zhang Y, Li SX, et al. Droplet-microfluidic-based promoter engineering and expression fine-tuning for improved erythromycin production in Saccharopolyspora erythraea NRRL 23338. Front Bioeng Biotechnol, 2022, 10: 864977. DOI:10.3389/fbioe.2022.864977
|
|
| [51] |
Ding RH, Hung KC, Mitra A, et al. Rapid isolation of antigen-specific B-cells using droplet microfluidics. RSC Adv, 2020, 10(45): 27006-27013. DOI:10.1039/D0RA04328A
|
|
| [52] |
Yuan HL, Zhou Y, Lin YP, et al. Microfluidic screening and genomic mutation identification for enhancing cellulase production in Pichia pastoris. Biotechnol Biofuels Bioprod, 2022, 15(1): 50. DOI:10.1186/s13068-022-02150-w
|
|
| [53] |
Yuan HL, Tu R, Tong XW, et al. Ultrahigh-throughput screening of industrial enzyme-producing strains by droplet-based microfluidic system. J Ind Microbiol Biotechnol, 2022, 49(3): 1-9temp.
|
|
| [54] |
Qiao YX, Zhao XY, Zhu J, et al. Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system. Lab Chip, 2017, 18(1): 190-196.
|
|
| [55] |
Tu R, Li LP, Yuan HL, et al. Biosensor-enabled droplet microfluidic system for the rapid screening of 3-dehydroshikimic acid produced in Escherichia coli. J Ind Microbiol Biotechnol, 2020, 47(12): 1155-1160. DOI:10.1007/s10295-020-02316-1
|
|
| [56] |
付首颖, 夏苗苗, 张祎凝, 等. 核黄素工业菌株高通量筛选方法的建立和应用. 生物技术通报, 2020, 36(4): 47-53. Fu SY, Xia MM, Zhang YN, et al. Establishment and application of high-throughput screening method of riboflavin industrial strain. Biotech Bull, 2020, 36(4): 47-53 (in Chinese).
|
|
| [57] | |
|
| [58] |
Hua EB, Zhang Y, Yun KY, et al. Whole-cell biosensor and producer co-cultivation-based microfludic platform for screening Saccharopolyspora erythraea with hyper erythromycin production. ACS Synth Biol, 2022, 11(8): 2697-2708. DOI:10.1021/acssynbio.2c00102
|
|
| [59] |
Gielen F, Hours R, Emond S, et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). PNAS, 2016, 113(47): E7383-E7389.
|
|
| [60] |
Wang XX, Ren LH, Su YT, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells. Anal Chem, 2017, 89(22): 12569-12577. DOI:10.1021/acs.analchem.7b03884
|
|
| [61] |
Wang XX, Xin Y, Ren LH, et al. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo. Sci Adv, 2020, 6(32): eabb3521. DOI:10.1126/sciadv.abb3521
|
|
| [62] |
Holland-Moritz DA, Wismer MK, Mann BF, et al. Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale. Angew Chem Int Ed Engl, 2020, 59(11): 4470-4477. DOI:10.1002/anie.201913203
|
|
| [63] |
Nitta N, Sugimura T, Isozaki A, et al. Intelligent image-activated cell sorting. Cell, 2018, 175(1): 266-276.e13.
|
|
| [64] |
Sesen M, Whyte G. Image-based single cell sorting automation in droplet microfluidics. Sci Rep, 2020, 10(1): 8736.
|
|
| [65] | |
|
| [66] |
Qi XN, Zhang YY, Tu R, et al. High-throughput screening and characterization of xylose-utilizing, ethanol-tolerant thermophilic bacteria for bioethanol production. J Appl Microbiol, 2011, 110(6): 1584-1591.
|
|
| [67] |
Sun L, Zhang H, Yuan HL, et al. A double-enzyme-coupled assay for high-throughput screening of succinic acid-producing strains. J Appl Microbiol, 2013, 114(6): 1696-1701.
|
|
| [68] |
Tu R, Lv T, Sun L, et al. Development of a simple colorimetric assay for determination of the isoamyl alcohol-producing strain. Appl Biochem Biotechnol, 2020, 192(2): 632-642.
|
|
| [69] |
Jiang D, Tu R, Bai P, et al. Directed evolution of cytochrome P450 for sterol epoxidation. Biotechnol Lett, 2013, 35(10): 1663-1668.
|
|
| [70] |
Lu XY, Liu YW, Yang YQ, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat Commun, 2019, 10(1): 1378.
|
|
| [71] |
Cai T, Sun HB, Qiao J, et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science, 2021, 373(6562): 1523-1527.
|
|
| [72] | |
|
| [73] |
Feng H, Yuan Y, Yang Z, et al. Genome-wide genotype-phenotype associations in microbes. J Biosci Bioeng, 2021, 132(1): 1-8.
|
|
| [74] |
Liu Y, Wang R, Liu J, et al. Base editor enables rational genome-scale functional screening for enhanced industrial phenotypes in Corynebacterium glutamicum. Sci Adv, 2022, 8(35): eabq2157.
|
|
| [75] |
Hanna RE, Hegde M, Fagre CR, et al. Massively parallel assessment of human variants with base editor screens. Cell, 2021, 184(4): 1064-1080.e20.
|
|
| [76] |
Cuella-Martin R, Hayward SB, Fan X, et al. Functional interrogation of DNA damage response variants with base editing screens. Cell, 2021, 184(4): 1081-1097.e19.
|
|
| [77] |
Despres PC, Dube AK, Seki M, et al. Perturbing proteomes at single residue resolution using base editing. Nat Commun, 2020, 11(1): 1871.
|
|
 2022, Vol. 38
2022, Vol. 38