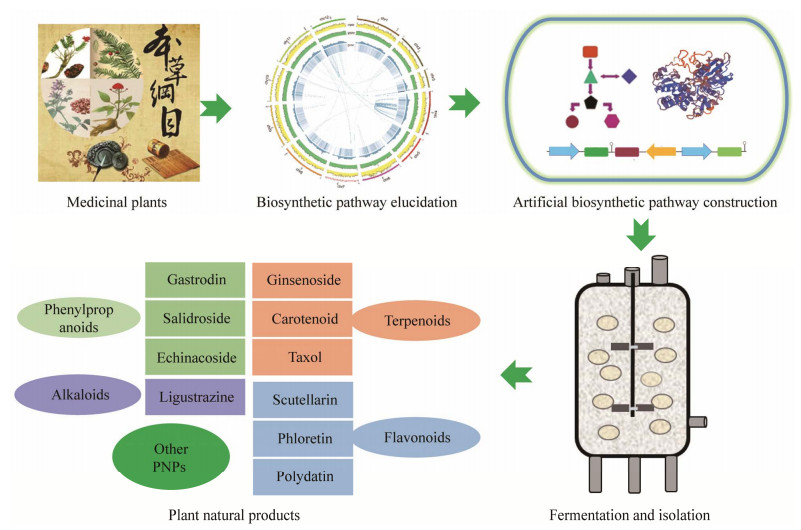
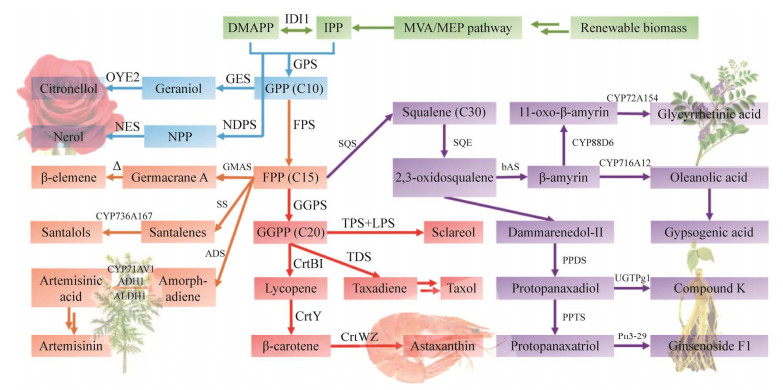
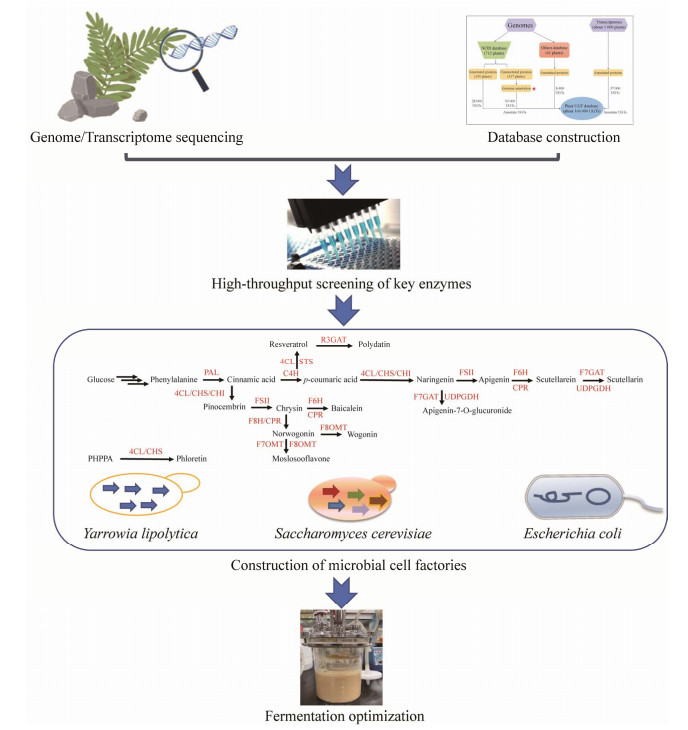
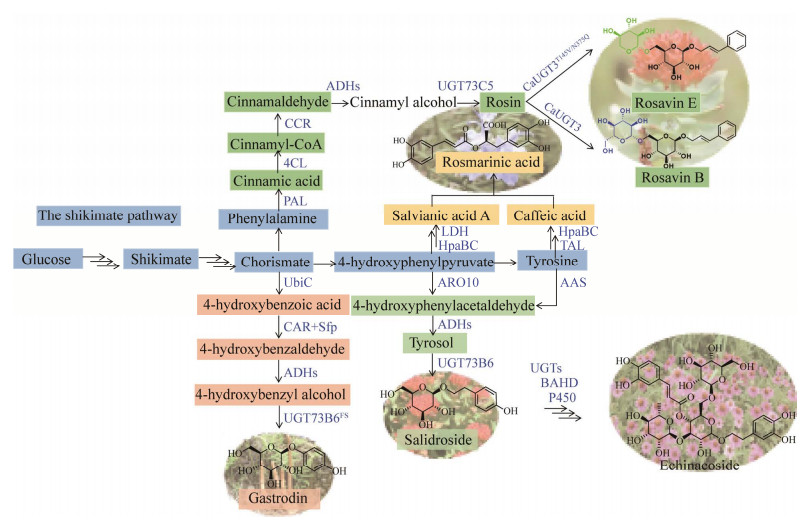
| [1] |
Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv, 2015, 33: 1582-1614. DOI:10.1016/j.biotechadv.2015.08.001
|
|
| [2] |
Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod, 2020, 83: 770-803. DOI:10.1021/acs.jnatprod.9b01285
|
|
| [3] |
David B, Wolfender J, Dias DA. The pharmaceutical industry and natural products: historical status and new trends. Phytochem Rev, 2015, 14: 299-315. DOI:10.1007/s11101-014-9367-z
|
|
| [4] |
Hidalgo D, Sanchez R, Lalaleo L, et al. Biotechnological production of pharmaceuticals and biopharmaceuticals in plant cell and organ cultures. Curr Med Chem, 2018, 25(30): 3577-3596. DOI:10.2174/0929867325666180309124317
|
|
| [5] |
Courdavault V, O'Connor SE, Jensen MK, et al. Metabolic engineering for plant natural products biosynthesis: new procedures, concrete achievements and remaining limits. Nat Prod Rep, 2021, 15: 2145-2153.
|
|
| [6] |
戴住波, 王勇, 周志华, 等. 植物天然产物合成生物学研究. 中国科学院院刊, 2018, 33(11): 1228-1238. Dai ZB, Wang Y, Zhou ZH, et al. Synthetic biology for production of plant-derived natural products. Bull Chin Acad Sci, 2018, 33(11): 1228-1238 (in Chinese). DOI:10.16418/j.issn.1000-3045.2018.11.011
|
|
| [7] |
Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature, 2013, 496: 528-532. DOI:10.1038/nature12051
|
|
| [8] |
Ajikumar PK, Xiao WH, Tyo KEJ, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science, 2010, 330: 70-74. DOI:10.1126/science.1191652
|
|
| [9] | |
|
| [10] |
Luo X, Reiter MA, D'Espaux L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature, 2019, 567: 123-126. DOI:10.1038/s41586-019-0978-9
|
|
| [11] |
Chang MC, Eachus RA, Trieu W, et al. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol, 2007, 3(5): 274-277. DOI:10.1038/nchembio875
|
|
| [12] |
王冬, 戴住波, 张学礼. 酵母人工合成细胞生产植物源天然产物. 微生物学报, 2016, 56(3): 14. Wang D, Dai ZB, Zhang XL. Production of plant-derived natural products in yeast cells. Acta Microbiol Sin, 2016, 56(3): 14 (in Chinese). DOI:10.13343/j.cnki.wsxb.20150416
|
|
| [13] |
Paddon CJ, Jiang H, Kung SH. Amorpha-4, 11-diene 12-mononzygenase variants and uses thereof (P). WO2021150960A1, 2021-07-29.
|
|
| [14] |
Zhou ZH, Wei W, Yan X, et al. Group of UDP-glycosyltransferase for catalyzing carbohydrate chain enlongation, and application thereof (P). WOCN18087678, 2018-05-21.
|
|
| [15] |
Zhou ZH, Yan X, Fan Y, et al. Group of glycosyltransferases and use thereof (P). WOCN2013088819, 2014-06-09.
|
|
| [16] |
Wang J, Li S, Xiong Z, et al. Pathway mining-based integration of critical enzyme parts for de novo biosynthesis of steviolglycosides sweetener in Escherichia coli. Cell Res, 2016, 26(2): 258-261. DOI:10.1038/cr.2015.111
|
|
| [17] |
Sun Y, Chen Z, Li J, et al. Diterpenoid UDP- glycosyltransferases from Chinese sweet tea and Ashitaba complete the biosynthesis of rubusoside. Mol Plant, 2018, 11(10): 1308-1311. DOI:10.1016/j.molp.2018.05.010
|
|
| [18] |
Ma T, Shi B, Ye Z, et al. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab Eng, 2019, 52: 134-142. DOI:10.1016/j.ymben.2018.11.009
|
|
| [19] |
张学礼, 黄璐琦, 戴住波, 等. 一种重组菌及其用途: PCT/CN2017/109029, 2017-11-02.
Zhang XL, Huang LQ, Dai ZB, et al. Recombinant yeast and use thereof: PCT/CN2017/109029, 2017-11-02 (in Chinese).
|
|
| [20] |
Li R, Wang K, Wang D, et al. Production of plant volatile terpenoids (rose oil) by yeast cell factories. Green Chem, 2021, 23(14): 5088-5096. DOI:10.1039/D1GC00917F
|
|
| [21] |
Lee SJ, Lee JS, Lee E, et al. The ginsenoside metabolite compound K inhibits hormone-independent breast cancer through downregulation of cyclin D1. Journal of Functional Foods, 2018, 46: 159-166. DOI:10.1016/j.jff.2018.04.050
|
|
| [22] |
Liao LM, Zhang Y, Lin SF, et al. In enzymatic transformation from protopanaxadiol ginsenoside Rb1 into rare ginsenoside C-K and its anti-cancer activity, 2nd International Conference on Biotechnology, Chemical and Materials Engineering (CBCME 2012), Xiamen, Peoples R China, Dec 28-29; Xiamen, Peoples R China, 2012; pp 752-755.
|
|
| [23] |
Lee KT, Jung TW, Lee HJ, et al. The antidiabetic effect of ginsenoside Rb2 via activation of AMPK. Archives of Pharmacal Research, 2011, 34(7): 1201-1208. DOI:10.1007/s12272-011-0719-6
|
|
| [24] |
Dai Z, Liu Y, Zhang X, et al. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng, 2013, 20: 146-156. DOI:10.1016/j.ymben.2013.10.004
|
|
| [25] |
王冬, 刘怡, 许骄阳, 等. 创建酿酒酵母细胞工厂高效生产人参皂苷前体达玛烯二醇Ⅱ. 药学学报, 2018, 53(8): 1233-1241. Wang D, Liu Y, Xu JY, et al. Construction of efficient yeast cell factories for production of ginsenosides precursor dammarenediol-Ⅱ. Acta Pharm Sin, 2018, 53(8): 1233-1241 (in Chinese). DOI:10.16438/j.0513-4870.2018-0503
|
|
| [26] |
Dai Z, Wang B, Liu Y, Shi M, et al. Producing aglycons of ginsenosides in bakers' yeast. Sci Rep, 2014, 4.
|
|
| [27] |
Wang D, Wang J, Shi Y, et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab Eng, 2020, 61: 131-140. DOI:10.1016/j.ymben.2020.05.007
|
|
| [28] |
Shi Y, Wang D, Li R, et al. Engineering yeast subcellular compartments for increased production of the lipophilic natural products ginsenosides. Metab Eng, 2021, 67: 104-111. DOI:10.1016/j.ymben.2021.06.002
|
|
| [29] |
Bai X, Zhang Y, Jiang H, et al. Effects of maslinic acid on the proliferation and apoptosis of A549 lung cancer cells. Mol Med Rep, 2016, 13(1): 117-122. DOI:10.3892/mmr.2015.4552
|
|
| [30] |
Miura T, Ueda N, Yamada K, Fukushima M, et al. Antidiabetic effects of corosolic acid in KK-Ay diabetic mice. Biol Pharm Bull, 2006, 29(3): 585-587. DOI:10.1248/bpb.29.585
|
|
| [31] |
Bonte F, Dumas M, Chaudagne C, et al. Influence of asiatic acid, madecassic acid, and asiaticoside on human collagen I synthesis. Planta Med, 1994, 60(2): 133-135. DOI:10.1055/s-2006-959434
|
|
| [32] |
Dai Z, Liu Y, Sun Z, et al. Identification of a novel cytochrome P450 enzyme that catalyzes the C-2α hydroxylation of pentacyclic triterpenoids and its application in yeast cell factories. Metab Eng, 2019, 51: 70-78. DOI:10.1016/j.ymben.2018.10.001
|
|
| [33] |
张学礼, 戴住波, 刘芸, 等. 三萜2位α-羟化酶MAA45及其相关生物材料与它们在制备山楂酸和科罗索酸中的应用. ZL201610236283.9, 2019-02-26.
Zhang XL, Dai ZB, Liu Y, et al. Triterpene 2 α-hydroxylase MAA45 and its related biomaterials and their application in the preparation of maslinic acid and corosolic acid: ZL201610236283.9, 2019-02-26 (in Chinese).
|
|
| [34] |
Li W, Ma X, Li G, et al. De novo biosynthesis of the oleanane-type triterpenoids of tunicosaponins in yeast. ACS Aynth Biol, 2021, 10(8): 1874-1881. DOI:10.1021/acssynbio.1c00065
|
|
| [35] |
Li Q, Fan F, Gao X, et al. Balanced activation of IspG and IspH to eliminate MEP intermediate accumulation and improve isoprenoids production in Escherichia coli. Metab Eng, 2017, 44: 13-21. DOI:10.1016/j.ymben.2017.08.005
|
|
| [36] |
Xie Q, Li S, Zhao D, et al. Manipulating the position of DNA expression cassettes using location tags fused to dCas9 (Cas9-Lag) to improve metabolic pathway efficiency. Microb Cell Fact, 2020, 19(1): 229. DOI:10.1186/s12934-020-01496-w
|
|
| [37] |
Li D, Li Y, Xu JY, et al. Engineering CrtW and CrtZ for improving biosynthesis of astaxanthin in Escherichia coli. Chin J Nat Med, 2020, 18(9): 666-676.
|
|
| [38] |
Zhang M, Gong Z, Tang J, et al. Improving astaxanthin production in Escherichia coli by co-utilizing CrtZ enzymes with different substrate preference. Microb Cell Fact, 2022, 21(1): 71. DOI:10.1186/s12934-022-01798-1
|
|
| [39] |
Zhao J, Li Q, Sun T, et al. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab Eng, 2013, 17: 42-50. DOI:10.1016/j.ymben.2013.02.002
|
|
| [40] |
Wu T, Ye L, Zhao D, et al. Membrane engineering-a novel strategy to enhance the production and accumulation of β-carotene in Escherichia coli. Metab Eng, 2017, 43(Pt A): 85-91.
|
|
| [41] |
Wu T, Li S, Ye L, et al. Engineering an artificial membrane vesicle trafficking system (AMVTS) for the excretion of β-carotene in Escherichia coli. ACS Synth Biol, 2019, 8(5): 1037-1046. DOI:10.1021/acssynbio.8b00472
|
|
| [42] |
Tong Y, Luo YF, Gao W. Biosynthesis of paclitaxel using synthetic biology. Phytochem Rev, 2021, 21: 863-877.
|
|
| [43] | |
|
| [44] |
Van Rijn JPM, Escorcia AM, Thiel W. QM/MM study of the taxadiene synthase mechanism. J Comput Chem, 2019, 40: 1902-1910. DOI:10.1002/jcc.25846
|
|
| [45] |
Li J, Mutanda I, Wang K, Yang L, et al. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat Commun, 2019, 10: 4850. DOI:10.1038/s41467-019-12879-y
|
|
| [46] |
Walls LE, Malci K, Nowrouzi B, et al. Optimizing the biosynthesis of oxygenated and acetylated taxol precursors in Saccharomyces cerevisiae using advanced bioprocessing strategies. Biotechnol Bioeng, 2021, 118: 279-293. DOI:10.1002/bit.27569
|
|
| [47] |
Edgar S, Zhou K, Qiao K, et al. Mechanistic insights into taxadiene epoxidation by taxadiene-5α-hydroxylase. ACS Chem Biol, 2016, 11: 460-469. DOI:10.1021/acschembio.5b00767
|
|
| [48] |
Biggs BW, Rouck JE, Kambalyal A, et al. Orthogonal assays clarify the oxidative biochemistry of taxol P450 CYP725A4. ACS Chem Biol, 2016, 11: 1445-1451. DOI:10.1021/acschembio.5b00968
|
|
| [49] |
Zhou K, Qiao K, Edgar S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol, 2015, 33: 377-383. DOI:10.1038/nbt.3095
|
|
| [50] |
Cheng J, Wang X, Liu X, et al. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol Plant, 2021, 14: 1199-1209. DOI:10.1016/j.molp.2021.04.015
|
|
| [51] |
Xiong X, Gou J, Liao Q, et al. The taxus genome provides insights into paclitaxel biosynthesis. Nat Plants, 2021, 7: 1026-1036. DOI:10.1038/s41477-021-00963-5
|
|
| [52] |
Song C, Fu F, Yang L, et al. Taxus yunnanensis genome offers insights into gymnosperm phylogeny and taxol production. Commun Biol, 2021, 4: 1203. DOI:10.1038/s42003-021-02697-8
|
|
| [53] |
Shen N, Wang T, Gan Q, et al. Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity. Food Chem, 2022, 383: 132531. DOI:10.1016/j.foodchem.2022.132531
|
|
| [54] |
Gomes D, Rodrigues LR, Rodrigues JL. Perspectives on the design of microbial cell factories to produce prenylflavonoids. Int J Food Microbiol, 2022, 367: 109588. DOI:10.1016/j.ijfoodmicro.2022.109588
|
|
| [55] |
Marsafari M, Samizadeh H, Rabiei B, et al. Biotechnological production of flavonoids: an update on plant metabolic engineering, microbial host selection, and genetically encoded biosensors. Biotechnol J, 2020, 15: e1900432. DOI:10.1002/biot.201900432
|
|
| [56] |
Shah FLA, Ramzi AB, Baharum SN, et al. Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol Biol Rep, 2019, 46: 6647-6659. DOI:10.1007/s11033-019-05066-1
|
|
| [57] |
Pandey RP, Parajuli P, Koffas MA, et al. Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv, 2016, 34: 634-662. DOI:10.1016/j.biotechadv.2016.02.012
|
|
| [58] |
Lou H, Hu L, Lu H, et al. Metabolic engineering of microbial cell factories for biosynthesis of flavonoids: a review. Molecules, 2021, 26: 4522-4538. DOI:10.3390/molecules26154522
|
|
| [59] |
Sun J, Sun W, Zhang G, et al. High efficient production of plant flavonoids by microbial cell factories: challenges and opportunities. Metab Eng, 2022, 70: 143-154. DOI:10.1016/j.ymben.2022.01.011
|
|
| [60] |
Li H, Lyv Y, Zhou S, et al. Microbial cell factories for the production of flavonoids-barriers and opportunities. Bioresour Technol, 2022, 360: 127538. DOI:10.1016/j.biortech.2022.127538
|
|
| [61] |
Zha J, Wu X, Gong G, et al. Pathway enzyme engineering for flavonoid production in recombinant microbes. Metab Eng Commun, 2019, 9: e00104. DOI:10.1016/j.mec.2019.e00104
|
|
| [62] |
Liu X, Cheng J, Zhang G, et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun, 2018, 9: 448. DOI:10.1038/s41467-018-02883-z
|
|
| [63] |
Wang Y, Liu X, Chen B, et al. Metabolic engineering of Yarrowia lipolytica for scutellarin production. Synthetic and Systems Biotechnology, 2022, 7: 958-964. DOI:10.1016/j.synbio.2022.05.009
|
|
| [64] |
Liu T, Liu Y, Li L, et al. De novo biosynthesis of polydatin in Saccharomyces cerevisiae. J Agric Food Chem, 2021, 69: 5917-5925. DOI:10.1021/acs.jafc.1c01557
|
|
| [65] |
Jiang C, Liu X, Chen X, et al. Raising the production of phloretin by alleviation of by-product of chalcone synthase in the engineered yeast. Sci China Life Sci, 2020, 63: 1734-1743. DOI:10.1007/s11427-019-1634-8
|
|
| [66] |
Duan L, Ding W, Liu X, et al. Biosynthesis and engineering of kaempferol in Saccharomyces cerevisiae. Microb Cell Fact, 2017, 16: 165. DOI:10.1186/s12934-017-0774-x
|
|
| [67] |
Liu X, Cheng J, Zhu X, et al. De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae. ACS Synth Biol, 2020, 9: 3042-3051. DOI:10.1021/acssynbio.0c00289
|
|
| [68] |
Bi H, Bai Y, Cai T, et al. Engineered short branched-chain acyl-CoA synthesis in E. coli and acylation of chloramphenicol to branched-chain derivatives. Appl Microbiol Biotechnol, 2013, 97(24): 10339-10348. DOI:10.1007/s00253-013-5262-6
|
|
| [69] |
Zhou W, Zhuang Y, Bai Y, et al. Biosynthesis of phlorisovalerophenone and 4-hydroxy-6-isobutyl-2- pyrone in Escherichia coli from glucose. Microb Cell Fact, 2016, 15(1): 149. DOI:10.1186/s12934-016-0549-9
|
|
| [70] |
Liu LQ, Liu H, Zhang W, et al. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering, 2019, 5(2): 287-295. DOI:10.1016/j.eng.2018.11.029
|
|
| [71] |
Liu Q, Yu T, Li X, et al. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat Commun, 2019, 10(1): 4976. DOI:10.1038/s41467-019-12961-5
|
|
| [72] |
Chen Z, Sun X, Li Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab Eng, 2017, 39: 102-109. DOI:10.1016/j.ymben.2016.10.021
|
|
| [73] |
Yao YF, Wang CS, Qiao J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab Eng, 2013, 19: 79-87. DOI:10.1016/j.ymben.2013.06.001
|
|
| [74] |
Li S, Liang C, Liu G, et al. De novo biosynthesis of chlorogenic acid using an artificial microbial community. J Agric Food Chem, 2021, 69(9): 2816-2825. DOI:10.1021/acs.jafc.0c07588
|
|
| [75] |
Babaei M, Borja Zamfir GM, Chen X, et al. Metabolic engineering of Saccharomyces cerevisiae for rosmarinic acid production. ACS Synth Biol, 2020, 9(8): 1978-1988. DOI:10.1021/acssynbio.0c00048
|
|
| [76] |
Schultz BJ, Kim SY, Lau W, et al. Total biosynthesis for milligram-scale production of etoposide intermediates in aplant chassis. J Am Chem Soc, 2019, 141(49): 19231-19235. DOI:10.1021/jacs.9b10717
|
|
| [77] | |
|
| [78] |
Yin H, Hu T, Zhuang Y, et al. Metabolic engineering of Saccharomyces cerevisiae for high-level production of gastrodin from glucose. Microb Cell Fact, 2022, 21(1): 18. DOI:10.1186/s12934-022-01747-y
|
|
| [79] |
Bai Y, Bi H, Zhuang Y, et al. Production of salidroside in metabolically engineered Escherichia coli. Sci Rep, 2014, 4: 6640.
|
|
| [80] |
Jiang J, Yin H, Wang S, et al. Metabolic engineering of Saccharomyces cerevisiae for high-level production of salidroside from glucose. J Agric Food Chem, 2018, 66(17): 4431-4438. DOI:10.1021/acs.jafc.8b01272
|
|
| [81] |
Torrens-Spence MP, Pluskal T, Li FS, et al. Complete pathway elucidation and heterologous reconstitution of Rhodiola salidroside biosynthesis. Mol Plant, 2018, 11(1): 205-217. DOI:10.1016/j.molp.2017.12.007
|
|
| [82] |
Chung D, Kim SY, Ahn JH. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli. Sci Rep, 2017, 7(1): 2578. DOI:10.1038/s41598-017-02042-2
|
|
| [83] |
Liu X, Li XB, Jiang JL, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng, 2018, 47: 243-253. DOI:10.1016/j.ymben.2018.03.016
|
|
| [84] |
Guo W, Huang Q, Feng Y, et al. Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae. Biotechnol Bioeng, 2020, 117(8): 2410-2419. DOI:10.1002/bit.27370
|
|
| [85] |
Liu H, Tian Y, Zhou Y, et al. Multi-modular engineering of Saccharomyces cerevisiae for high-titre production of tyrosol and salidroside. Microb Biotechnol, 2021, 14(6): 2605-2616. DOI:10.1111/1751-7915.13667
|
|
| [86] |
Li X, Zhou Z, Li W, et al. Design of stable and self-regulated microbial consortia for chemical synthesis. Nat Commun, 2022, 13(1): 1554. DOI:10.1038/s41467-022-29215-6
|
|
| [87] |
蔡岩, 苗志伟. 玫瑰红景天药用历史及功能简介. 化学通报, 2021, 84: 1108-1119. Cai Y, Miao ZW. Introduction to the medicinal history and functions of Rhodiola Rosea. Chemistry, 2021, 84: 1108-1119 (in Chinese). DOI:10.14159/j.cnki.0441-3776.2021.10.015
|
|
| [88] |
Patov SA, Punegov VV, Kuchin AV. Synthesis of the Rhodiola rosea glycoside rosavin. Chem Nat Compd, 2006, 42: 397-399. DOI:10.1007/s10600-006-0165-8
|
|
| [89] |
Zhou W, Bi H, Zhuang Y, et al. Production of cinnamyl alcohol glucoside from glucose in Escherichia coli. J Agric Food Chem, 2017, 65: 2129-2135. DOI:10.1021/acs.jafc.7b00076
|
|
| [90] |
Bi H, Wang S, Zhou W, et al. Producing Gram-scale unnatural rosavin analogues from glucose by engineered Escherichia coli. ACS Synth Biol, 2019, 8: 1931-1940. DOI:10.1021/acssynbio.9b00219
|
|
| [91] |
Bi H, Qu G, Wang S, et al. Biosynthesis of a rosavin natural product in Escherichia coli by glycosyltransferase rational design and artificial pathway construction. Metab Eng, 2022, 69: 15-25. DOI:10.1016/j.ymben.2021.10.010
|
|
| [92] |
Swamy MK, Sinniah UR, Ghasemzadeh A. Anticancer potential of rosmarinic acid and its improved production through biotechnological interventions and functional genomics. Appl Microbiol Biotechnol, 2018, 102(18): 7775-7793. DOI:10.1007/s00253-018-9223-y
|
|
| [93] |
Khojasteh A, Mirjalili MH, Alcalde MA, et al. Powerful plant antioxidants: a new biosustainable approach to the production of rosmarinic acid. Antioxidants (Basel), 2020, 9(12): 1273. DOI:10.3390/antiox9121273
|
|
| [94] |
Jiang J, Bi H, Zhuang Y, et al. Engineered synthesis of rosmarinic acid in Escherichia coli resulting production of a new intermediate, caffeoyl- phenyllactate. Biotechnol Lett, 2016, 38(1): 81-88. DOI:10.1007/s10529-015-1945-7
|
|
| [95] |
Zhuang Y, Jiang J, Bi H, et al. Synthesis of rosmarinic acid analogues in Escherichia coli. Biotechnol Lett, 2016, 38(4): 619-627. DOI:10.1007/s10529-015-2011-1
|
|
| [96] |
Levsh O, Pluskal T, Carballo V, et al. Independent evolution of rosmarinic acid biosynthesis in two sister families under the lamiids clade of flowering plants. J Biol Chem, 2019, 294(42): 15193-15205. DOI:10.1074/jbc.RA119.010454
|
|
| [97] |
Yang Y, Wu Y, Zhuang Y, et al. Discovery of glycosyltransferases involved in the biosynthesis of ligupurpuroside B. Org Lett, 2021, 23(20): 7851-7854. DOI:10.1021/acs.orglett.1c02873
|
|
| [98] | |
|
| [99] |
Galanie S, Thodey K, Trenchard IJ, et al. Complete biosynthesis of opioids in yeast. Science, 2015, 349(6252): 1095-1100. DOI:10.1126/science.aac9373
|
|
| [100] |
Qiu F, Yang C, Yuan L, et al. A phenylpyruvic acid reductase is required for biosynthesis of tropane alkaloids. Org Lett, 2018, 20(24): 7807-7810. DOI:10.1021/acs.orglett.8b03236
|
|
| [101] |
Qiu F, Zeng J, Wang J, et al. Functional genomics analysis reveals two novel genes required for littorine biosynthesis. New Phytol, 2020, 225(5): 1906-1914. DOI:10.1111/nph.16317
|
|
| [102] |
Fei Qiu, Yijun Yan, Junlan Zeng, et al. Biochemical and mtabolic insights into hyoscyamine dehydrogenase. ACS Catalysis, 2021, 11(5): 2912-2924. DOI:10.1021/acscatal.0c04667
|
|
| [103] |
Peng K, Guo D, Lou Q, et al. Synthesis of ligustrazine from acetaldehyde by a combined biological-chemical approach. ACS Synth Biol, 2020, 9: 2902-2908. DOI:10.1021/acssynbio.0c00113
|
|
| [104] |
Liu X, Zhu X, Wang H, et al. Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synth Syst Biotechnol, 2020, 5: 187-199. DOI:10.1016/j.synbio.2020.06.008
|
|
| [105] |
Wang H, Wang Q, Liu Y, Liao X, Chu H, Chang H, Cao Y, Li Z, Zhang T, Cheng J, Jiang H. PCPD: plant cytochrome P450 database and web-based tools for structural construction and ligand docking. Synth Syst Biotechnol, 2021, 2: 102-109.
|
|
| [106] |
Ding W, Jiang H. Engineering microbial cell factories for the production of plant natural products: from design principles to industrial-scale production. Microb Cell Fact, 2017, 16: 125-133. DOI:10.1186/s12934-017-0732-7
|
|
| [107] |
张震, 曾雪城, 秦磊, 等. 微生物细胞工厂的智能设计进展. 化工学报, 2021, 72(12): 6093-6108. Zhang Z, Zeng XC, Qin L, et al. Intelligent design of microbial cell factory. CIESC J, 2021, 72(12): 6093-6108 (in Chinese).
|
|
| [108] |
Ding S, Tian Y, Cai P, et al. novoPathFinder: a webserver of designing novel-pathway with integrating GEM-model. Nucleic Acids Res, 2020, 48(W1): W477-W487. DOI:10.1093/nar/gkaa230
|
|
| [109] |
Hadadi N, MohammadiPeyhani H, Miskovic L, et al. Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites. PNAS, 2019, 116(15): 7298-7307. DOI:10.1073/pnas.1818877116
|
|
| [110] |
Mahood EH, Kruse LH, Moghe GD. Machine learning: a powerful tool for gene function prediction in plants. Appl Plant Sci., 2020, 8(7): e11376.
|
|
| [111] |
Zhu X, Liu X, Liu T, Wang Y, et al. Synthetic biology of plant natural products: from pathway elucidation to engineered biosynthesis in plant cells. Plant Commun, 2021, 2: 100229. DOI:10.1016/j.xplc.2021.100229
|
|
| [112] |
魏笑莲, 钱智玲, 陈巧巧, 等. 遗传改造微生物制造食品和饲料的监管要求及欧盟授权案例分析. 合成生物学, 2021, 2(1): 121-133. Wei XL, Qian ZL, Chen QQ, et al. Regulatory requirements for food and feed produced with genetically modified microorganisms. Syn Bio J, 2021, 2(1): 121-133 (in Chinese).
|
|
 2022, Vol. 38
2022, Vol. 38