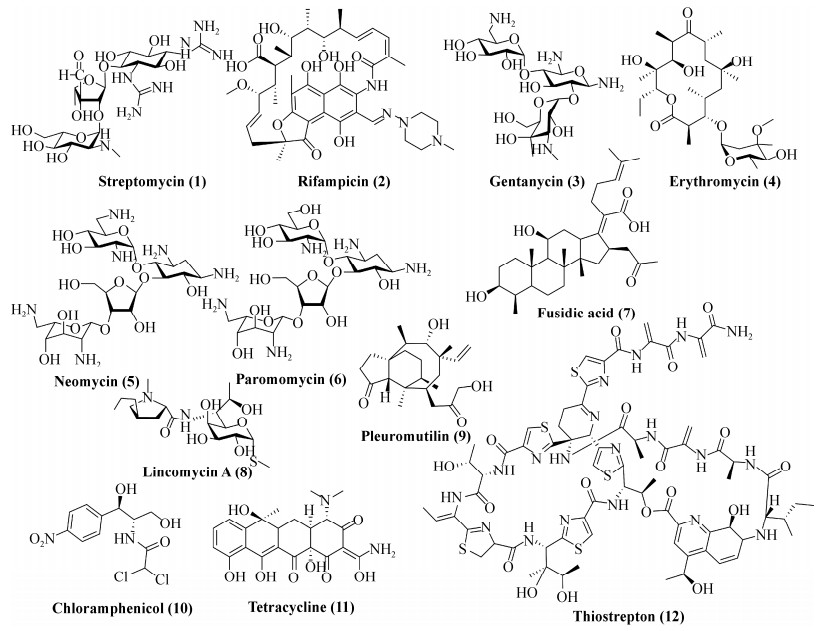
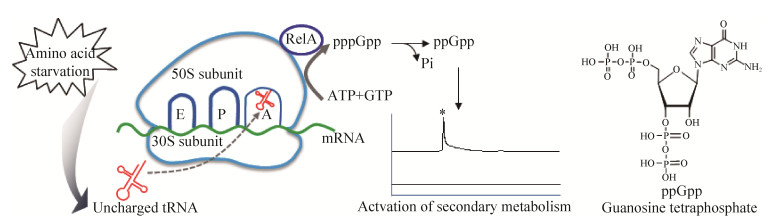
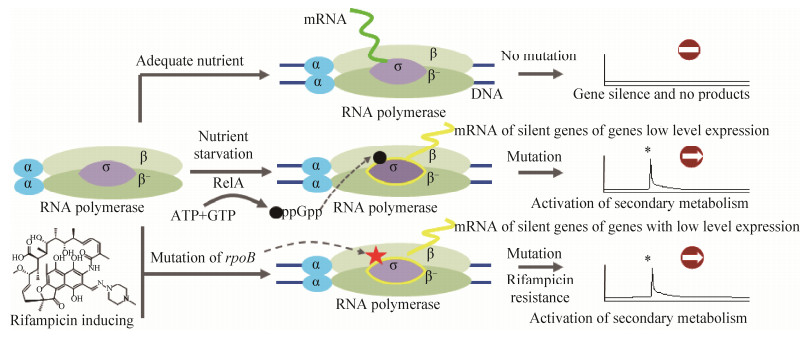
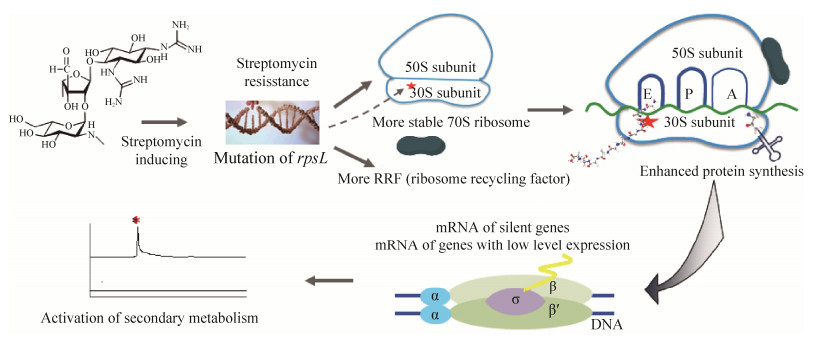
| [1] | |
|
| [2] |
Genilloud O. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep, 2017, 34(10): 1203-1232. DOI:10.1039/C7NP00026J
|
|
| [3] |
Nepal KK, Wang G. Streptomycetes: surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products. Biotechnol Adv, 2019, 37(1): 1-20. DOI:10.1016/j.biotechadv.2018.10.003
|
|
| [4] |
Liu R, Deng Z, Liu T. Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab Eng, 2018, 50: 74-84. DOI:10.1016/j.ymben.2018.05.015
|
|
| [5] |
Palazzotto E, Tong Y, Lee SY, et al. Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv, 2019, 37(6): 107366. DOI:10.1016/j.biotechadv.2019.03.005
|
|
| [6] | |
|
| [7] |
Hug JJ, Bader CD, Remškar M, et al. Concepts and methods to access novel antibiotics from actinomycetes. Antibiotics (Basel), 2018, 7(2): 44. DOI:10.3390/antibiotics7020044
|
|
| [8] | |
|
| [9] | |
|
| [10] | |
|
| [11] |
Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol, 2015, 13(8): 509-523. DOI:10.1038/nrmicro3496
|
|
| [12] |
Ochi K. Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J Antibiot (Tokyo), 2017, 70(1): 25-40. DOI:10.1038/ja.2016.82
|
|
| [13] | |
|
| [14] |
Ochi K. From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem, 2007, 71(6): 1373-1386. DOI:10.1271/bbb.70007
|
|
| [15] |
Hosaka T, Ohnishi-Kameyama M, Muramatsu H, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol, 2009, 27(5): 462-464. DOI:10.1038/nbt.1538
|
|
| [16] |
Ochi K, Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol, 2013, 97(1): 87-98. DOI:10.1007/s00253-012-4551-9
|
|
| [17] |
Mao D, Okada BK, Wu Y, et al. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol, 2018, 45: 156-163. DOI:10.1016/j.mib.2018.05.001
|
|
| [18] |
Baral B, Akhgari A, Metsä-Ketelä M. Activation of microbial secondary metabolic pathways: avenues and challenges. Synth Syst Biotechnol, 2018, 3(3): 163-178. DOI:10.1016/j.synbio.2018.09.001
|
|
| [19] |
Zhu S, Duan Y, Huang Y. The Application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiotics (Basel), 2019, 8(3): 133. DOI:10.3390/antibiotics8030133
|
|
| [20] |
Tan GY, Liu T. Rational synthetic pathway refactoring of natural products biosynthesis in actinobacteria. Metab Eng, 2017, 39: 228-236. DOI:10.1016/j.ymben.2016.12.006
|
|
| [21] | |
|
| [22] |
Irving SE, Corrigan RM. Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology, 2018, 164(3): 268. DOI:10.1099/mic.0.000621
|
|
| [23] |
Artsimovitch I, Patlan V, Sekine S, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell, 2004, 117(3): 299-310. DOI:10.1016/S0092-8674(04)00401-5
|
|
| [24] |
Okamoto S, Lezhava A, Hosaka T, et al. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in S treptomyces coelicolor A3(2). J Bacteriol, 2003, 185(2): 601-609. DOI:10.1128/JB.185.2.601-609.2003
|
|
| [25] |
Thong WL, Shin-ya K, Nishiyama M, et al. Discovery of an antibacterial isoindolinone-containing tetracyclic polyketide by cryptic gene activation and characterization of its biosynthetic gene cluster. ACS Chem Biol, 2018, 13(9): 2615-2622. DOI:10.1021/acschembio.8b00553
|
|
| [26] |
Thong WL, Shin-ya K, Nishiyama M, et al. Methylbenzene-containing polyketides from a Streptomyces that spontaneously acquired rifampicin resistance: structural elucidation and biosynthesis. J Nat Prod, 2016, 79(4): 857-864. DOI:10.1021/acs.jnatprod.5b00922
|
|
| [27] |
Tanaka Y, Kasahara K, Hirose Y, et al. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance ( rpoB) mutations in actinomycetes. J Bacteriol, 2013, 195(13): 2959-2970. DOI:10.1128/JB.00147-13
|
|
| [28] |
Ma Z, Luo S, Xu XH, et al. Characterization of representative rpoB gene mutations leading to a significant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J Ind Microbiol Biotechnol, 2016, 43(4): 463-471. DOI:10.1007/s10295-015-1732-4
|
|
| [29] |
Shima J, Hesketh A, Okamoto S, et al. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol, 1996, 178(24): 7276-7284. DOI:10.1128/jb.178.24.7276-7284.1996
|
|
| [30] |
Hesketh A, Ochi K. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J Antibiot, 1997, 50(6): 532-535. DOI:10.7164/antibiotics.50.532
|
|
| [31] |
Ochi K, Zhang D, Kawamoto S, et al. Molecular and functional analysis of the ribosomal L11 and S12 protein genes ( rplK and rpsL) of Streptomyces coelicolor A3(2). Mol Gen Genet, 1997, 256(5): 488-498.
|
|
| [32] |
Okamoto-Hosoya Y, Sato T, Ochi K. Resistance to paromomycin is conferred by rpsL mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2). J Antibiot, 2000, 53(12): 1424-1427. DOI:10.7164/antibiotics.53.1424
|
|
| [33] |
Hu H, Ochi K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl Environ Microbiol, 2001, 67(4): 1885-1892. DOI:10.1128/AEM.67.4.1885-1892.2001
|
|
| [34] |
Xu J, Tozawa Y, Lai C, et al. Rifampicin resistant mutation in the rpoB gene confers ppGpp-independent antibiotic productivity to Streptomyces coelicolor and Streptomyces griseus. Mol Gen Genet, 2002, 268(2): 179-189. DOI:10.1007/s00438-002-0730-1
|
|
| [35] |
Hosaka T, Xu J, Ochi K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol Microbiol, 2006, 61(4): 883-897. DOI:10.1111/j.1365-2958.2006.05285.x
|
|
| [36] |
Nishimura K, Hosaka T, Tokuyama S, et al. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J Bacteriol, 2007, 189(10): 3876-3883. DOI:10.1128/JB.01776-06
|
|
| [37] |
Wang G, Hosaka T, Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol, 2008, 74(9): 2834-2840. DOI:10.1128/AEM.02800-07
|
|
| [38] |
Tanaka Y, Komatsu M, Okamoto S, et al. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol, 2009, 75(14): 4919-4922. DOI:10.1128/AEM.00681-09
|
|
| [39] |
Wang G, Inaoka T, Okamoto S, et al. A novel insertion mutation in Streptomyces coelicolor ribosomal S12 protein results in paromomycin resistance and antibiotic overproduction. Antimicrob Agents Chemother, 2009, 53(3): 1019-1026. DOI:10.1128/AAC.00388-08
|
|
| [40] |
Imai Y, Fujiwara T, Ochi K, et al. Development of the ability to produce secondary metabolites in Streptomyces through the acquisition of erythromycin resistance. J Antibiot, 2012, 65(6): 323-326. DOI:10.1038/ja.2012.16
|
|
| [41] | |
|
| [42] |
Hu H, Zhang Q, Ochi K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J Bacteriol, 2002, 184(14): 3984-3991. DOI:10.1128/JB.184.14.3984-3991.2002
|
|
| [43] |
Okamoto-Hosoya Y, Okamoto S, Ochi K. Development of antibiotic-overproducing strains by site-directed mutagenesis of the rpsL gene in Streptomyces lividans. Appl Environ Microbiol, 2003, 69(7): 4256-4259. DOI:10.1128/AEM.69.7.4256-4259.2003
|
|
| [44] |
Hosoya Y, Okamoto S, Muramatsu H, et al. Acquisition of certain streptomycin-resistant ( str) mutations enhances antibiotic production in bacteria. Antimicrob Agents Chemother, 1998, 42(8): 2041-2047. DOI:10.1128/AAC.42.8.2041
|
|
| [45] | |
|
| [46] |
Li L, Guo J, Wen Y, et al. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol, 2010, 37(7): 673-679. DOI:10.1007/s10295-010-0710-0
|
|
| [47] |
Lv XA, Jin YY, Li YD, et al. Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl Microbiol Biotechnol, 2013, 97(2): 641-648. DOI:10.1007/s00253-012-4322-7
|
|
| [48] |
Wang L, Zhao Y, Liu Q, et al. Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J Appl Microbiol, 2012, 112(6): 1095-1101. DOI:10.1111/j.1365-2672.2012.05302.x
|
|
| [49] |
Xie Y, Chen J, Wang B, et al. Activation and enhancement of caerulomycin A biosynthesis in marine-derived Actinoalloteichus sp. AHMU CJ021 by combinatorial genome mining strategies. Microb Cell Fact, 2020, 19(1): 159. DOI:10.1186/s12934-020-01418-w
|
|
| [50] |
Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol, 2011, 4(2): 207. DOI:10.1111/j.1751-7915.2010.00219.x
|
|
| [51] |
齐欢, 马正, 薛正莲, 等. 利福霉素抗性突变菌 Streptomyces sp.HS-NF-1046R中的两个新化合物. 药学学报, 2019, 54(1): 117-121. Qi H, Ma Z, Xue ZL, et al. Two new compounds from rifamycin resistant mutant strain Streptomyces sp. HS-NF-1046R. Acta Pharm Sin, 2019, 54(1): 117-121 (in Chinese).
|
|
| [52] |
Li LL, Ma TM, Liu Q, et al. Improvement of daptomycin production in Streptomyces roseosporus through the acquisition of pleuromutilin resistance. Biomed Res Int, 2013, 2013: 479742.
|
|
| [53] |
Yu G, Hui M, Li R, et al. Enhancement of daptomycin production by the method of combining ribosome engineering and genome shuffling in Streptomyces roseosporus. Appl Biochem Microbiol, 2018, 54(6): 611-615. DOI:10.1134/S0003683818060169
|
|
| [54] |
Zhang Y, Huang H, Xu S, et al. Activation and enhancement of fredericamycin A production in deepsea-derived Streptomyces somaliensis SCSIO ZH66 by using ribosome engineering and response surface methodology. Microb Cell Fact, 2015, 14: 64. DOI:10.1186/s12934-015-0244-2
|
|
| [55] |
Beltrametti F, Rossi R, Selva E, et al. Antibiotic production improvement in the rare actinomycete Planobispora rosea by selection of mutants resistant to the aminoglycosides streptomycin and gentamycin and to rifamycin. J Ind Microbiol Biotechnol, 2006, 33(4): 283-288. DOI:10.1007/s10295-005-0061-4
|
|
| [56] |
Fu P, Jamison M, La S, et al. Inducamides A-C, chlorinated alkaloids from an RNA polymerase mutant strain of Streptomyces sp.. Org Lett, 2014, 16(21): 5656-5659. DOI:10.1021/ol502731p
|
|
| [57] |
Wang XJ, Wang XC, Xiang WS. Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet- and chemically induced mutants. World J Microbiol Biotechnol, 2009, 25(6): 1051-1056. DOI:10.1007/s11274-009-9986-5
|
|
| [58] |
Derewacz DK, Goodwin CR, McNees CR, et al. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. PNAS, 2013, 110(6): 2336-2341. DOI:10.1073/pnas.1218524110
|
|
| [59] |
王耀耀, 刘云清, 朱研研, 等. 组合链霉素和利福平抗性突变去甲基万古霉素高产菌株的选育. 中国抗生素杂志, 2006, 31(4): 243-246. Wang YY, Liu YQ, Zhu YY, et al. Screening of high yield norvancomycin producing strain by streptomycin and rifampicin resistant mutation. Chin J Antibio, 2006, 31(4): 243-246 (in Chinese). DOI:10.3969/j.issn.1001-8689.2006.04.014
|
|
| [60] |
Wang Q, Zhang D, Li Y, et al. Genome shuffling and ribosome engineering of Streptomyces actuosus for high-yield nosiheptide production. Appl Biochem Biotechnol, 2014, 173(6): 1553-1563. DOI:10.1007/s12010-014-0948-5
|
|
| [61] |
Li YM, Li JY, Ye ZM, et al. Enhancement of angucycline production by combined UV mutagenesis and ribosome engineering and fermentation optimization in Streptomyces dengpaensis XZHG99 T. Prep Biochem Biotechnol, 2021, 51(2): 173-182. DOI:10.1080/10826068.2020.1805754
|
|
| [62] |
Zhao YF, Song ZQ, Ma Z, et al. Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J Ind Microbiol Biotechnol, 2019, 46(5): 697-708. DOI:10.1007/s10295-019-02146-w
|
|
| [63] |
Tamehiro N, Hosaka T, Xu J, et al. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl Environ Microbiol, 2003, 69(11): 6412-6417. DOI:10.1128/AEM.69.11.6412-6417.2003
|
|
| [64] |
Zhang KP, Mohsin A, Dai YC, et al. Combinatorial effect of ARTP mutagenesis and ribosome engineering on an industrial strain of Streptomyces albus S12 for enhanced biosynthesis of salinomycin. Front Bioeng Biotechnol, 2019, 7: 212. DOI:10.3389/fbioe.2019.00212
|
|
| [65] |
Li D, Zhang JH, Tian YQ, et al. Enhancement of salinomycin production by ribosome engineering in Streptomyces albus. Sci China Life Sci, 2019, 62(2): 276-279. DOI:10.1007/s11427-018-9474-7
|
|
| [66] |
Li D, Tian YQ, Liu X, et al. Reconstitution of a mini-gene cluster combined with ribosome engineering led to effective enhancement of salinomycin production in Streptomyces albus. Microb Biotechno, 2020.
|
|
| [67] |
Fukuda K, Tamura T, Ito H, et al. Production improvement of antifungal, antitrypanosomal nucleoside sinefungin by rpoB mutation and optimization of resting cell system of S treptomyces incarnatus NRRL 8089. J Biosci Bioeng, 2010, 109(5): 459-465. DOI:10.1016/j.jbiosc.2009.10.017
|
|
| [68] |
Liu L, Pan J, Wang Z, et al. Ribosome engineering and fermentation optimization leads to overproduction of tiancimycin A, a new enediyne natural product from Streptomyces sp. CB03234. J Ind Microbiol Biotechnol, 2018, 45(3): 141-151. DOI:10.1007/s10295-018-2014-8
|
|
| [69] |
Zhuang ZK, Jiang CZ, Zhang F, et al. Streptomycin-induced ribosome engineering complemented with fermentation optimization for enhanced production of 10-membered enediynes tiancimycin-A and tiancimycin-D. Biotechnol Bioeng, 2019, 116(6): 1304-1314. DOI:10.1002/bit.26944
|
|
| [70] |
Liu H, Jiang C, Lin J, et al. Genome shuffling based on different types of ribosome engineering mutants for enhanced production of 10-membered enediyne tiancimycin-A. Appl Microbiol Biotechnol, 2020, 104(10): 4359-4369. DOI:10.1007/s00253-020-10583-2
|
|
| [71] |
Ma Z, Tao LB, Bechthold A, et al. Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol, 2014, 98(11): 5051-5058. DOI:10.1007/s00253-014-5573-2
|
|
| [72] |
Tong QQ, Zhou YH, Chen XS, et al. Genome shuffling and ribosome engineering of Streptomyces virginiae for improved virginiamycin production. Bioprocess Biosyst Eng, 2018, 41(5): 729-738. DOI:10.1007/s00449-018-1906-3
|
|
| [73] |
Zhu X, Kong J, Yang H, et al. Strain improvement by combined UV mutagenesis and ribosome engineering and subsequent fermentation optimization for enhanced 6'-deoxy-bleomycin Z production. Appl Microbiol Biotechnol, 2018, 102(4): 1651-1661. DOI:10.1007/s00253-017-8705-7
|
|
| [74] |
Mukai K, Kobayashi M, Hoshino K, et al. Lincomycin-induced secondary metabolism in Streptomyces lividans 66 with a mutation in the gene encoding the RNA polymerase beta subunit. Curr Microbiol, 2020, 77(10): 2933-2939. DOI:10.1007/s00284-020-02126-9
|
|
| [75] |
Koshla O, Lopatniuk M, Borys O, et al. Genetically engineered rpsL merodiploidy impacts secondary metabolism and antibiotic resistance in Streptomyces. World J Microbiol Biotechnol, 2021, 37(4): 62. DOI:10.1007/s11274-021-03030-5
|
|
 2022, Vol. 38
2022, Vol. 38