中国科学院微生物研究所、中国微生物学会主办
文章信息
- 常珍珍, 龚桂芝, 彭祝春, 杨程, 洪棋斌
- CHANG Zhenzhen, GONG Guizhi, PENG Zhuchun, YANG Cheng, HONG Qibin
- 植物环状RNA研究进展
- Progress in circular RNAs of plants
- 生物工程学报, 2022, 38(5): 1706-1723
- Chinese Journal of Biotechnology, 2022, 38(5): 1706-1723
- 10.13345/j.cjb.210823
-
文章历史
- Received: November 4, 2021
- Accepted: December 21, 2021
- Published: December 30, 2021
非编码RNA (non-coding RNAs, ncRNAs),包括microRNA (miRNA)、小干扰RNA (small interfering RNA, siRNA)、环状RNA (circular RNA, circRNA) 和长链非编码RNA (long non-coding RNA, lncRNA),在真核生物的转录组序列中占很大比例,参与蛋白编码基因在转录和转录后水平上的表达调控[1-3]。circRNA是近年来加入非编码RNA家族的新成员,具有共价闭合的环状结构,区别于大多数线性RNA。研究表明,circRNA结构稳定、种类丰富、序列保守、在细胞和组织中特异性表达,并具备很多潜在功能,在调控基因表达和各种生物学功能方面发挥着重要作用[4]。高通量测序结果显示,circRNA广泛存在于多种植物中,参与花的发育、果实的成熟和胁迫响应等生长发育进程[5-6]。本文从circRNA的发现引入,总结了植物circRNA的鉴定工具、特征、主要类型和生成机制;结合动物中的研究,概述了植物circRNA的潜在功能、降解和定位,对植物circRNA研究存在的问题进行了讨论,并对进一步研究作了展望,期望能为植物circRNA的研究提供参考。
1 circRNA的发现1976年,Sanger等首次在植物类病毒中发现了闭合环状的RNA分子[7],紧接着,1980年,Arnberg等在酵母(Saccharomyces cerevisiae H.) 线粒体中发现类似的环状转录本[8],1986年Kos等发现了第一个含有circRNA的动物病毒-丁型肝炎病毒(hepatitis delta virus, HDV)[9],1990年Matsumoto等首次在真核细胞中发现circRNA的身影[10]。1991年Nigro等在对人类结肠癌缺失(delete in colorectal carcinoma, DCC) 细胞转录本的研究中,首次发现circRNA来源于内源RNA[11]。此后,研究人员在多种生物中均发现了circRNA,包括小鼠(Mus musculus)[12]、斑马鱼(Danio rerio)[13]、秀丽隐杆线虫(Caenorhabditis elegans, C. elegans)[14]等,甚至古菌(archaea)[15]中也发现了circRNA的身影。目前已经在近30种植物中发现了大量circRNA,包括拟南芥(Arabidopsis thaliana)[5]、水稻(Oryza sativa L.)[6]、大麦(Hordeum vulgare L.)[16]、番茄(Lycopersicon esculentum Miller)[17]、大豆(Glycine max (Linn.) Merr.)[18]、小麦(Triticum aestivum L.)[19]、玉米(Zea mays L.)[20]、猕猴桃(Actinidia)[21]、葡萄(Vitis vinifera Linn.)[22]和杜梨(Pyrus betulifolia Bunge)[23]以及中草药[24]等。
虽然早在几十年前就已发现circRNA,但由于circRNA不具有游离的3′和5′末端,无法通过快速扩增cDNA末端(rapid amplification of cDNA ends, RACE) 或基于poly(A) RNA建库的二代测序技术实现检测[25-26];同时,可环化外显子是经反向剪接连接的,异于经典的线性剪接,早期转录组分析的映射算法无法直接将测序得到的片段匹配到基因组,使得人们一度认为circRNA只是错误剪接的副产物[27]。随着高通量测序技术和生物信息学工具的发展,2012年Salzman等首次发现circRNA是由前体mRNA (pre-mRNA) 经反向剪接产生的环状分子,并且大量存在于人类不同类型的细胞中[28];2015年,Ye等在水稻和拟南芥中进行了植物circRNA的全基因组鉴定,在水稻根茎组织中鉴定到12 037个circRNA,拟南芥叶片中鉴定到6 012个circRNA[29],引发了植物circRNA研究的热潮。
2 植物circRNA的鉴定工具目前研究人员已经开发出许多计算工具来预测circRNA (表 1,在刘旭庆等[60]、Chen等[61]基础上补充),例如pcircRNA_finder[57]是唯一为植物circRNA预测而设计的软件,它结合了多个算法来检测反向剪接读数,为植物circRNA提供了一种更全面、更灵敏、更精确的预测方法,但是它仅能鉴定外显子circRNA;CircPlant[58]考虑植物和哺乳动物基因组的差异,应用几种特定于植物的标准从RNA-seq数据中准确地检测植物circRNA并预测其功能,特异性、准确度和预测效率更优。但是这些生物信息学工具在识别circRNA时,精确度、灵敏度和计算成本方面表现不同,因此使用不同的软件进行同时预测分析可以提高circRNA鉴定的效率。
| Tool names | Uniform resource locator | Description | References |
| CircPro | http://bis.zju.edu.cn/CircPro]]> | Detection of circRNAs with protein-coding potential. | [30] |
| CIRCfinder | https://github.com/YangLab/CIRCfinder]]> | Using junction reads to identify circular intronic RNAs. | [31] |
| Acfs | https://code.google.com/p/acfs/]]> | Using single- and paired-ended RNA-Seq data to identify and quantify circRNAs. | [32] |
| CircRNAFisher | https://github.com/duolinwang/CircRNAFisher]]> | Using different back splicing junction reads to identify circRNAs. | [33] |
| Circtools | https://github.com/dieterich-lab/circtools]]> | Providing a complete workflow for circRNAs from prediction to functional insights. | [34] |
| FUCHS | https://github.com/dieterich-lab/FUCHS]]> | Long reads data based to learn more about the exon coverage, the number of double break point fragments and the alternatively spliced exons. | [35] |
| PRAPI | http://www.bioinfor.org/bioinfor/tool/PRAPI/]]> | ISO-seq data based to analyze alternative transcription initiation, alternative splicing and circRNAs. | [36] |
| CircView | https://github.com/GeneFeng/CircView]]> | Help to understand potential functions of circRNAs and design the experiments. | [37] |
| CircPrimer | http://www.bioinf.com.cn/]]> | Allow to search, annotate, and visualize circRNAs, help to design primers and to determine the specificity of the primers. | [38] |
| Circseq_cup | https://github.com/bioinplant/circseq-cup/]]> | Using the back splicing RNA-Seq and paired-end reads to assemble the full-length sequences of circRNAs. | [39] |
| CIRI-full | https://sourceforge.net/projects/ciri-full/]]> | An approach for reconstruction of full-length circRNAs and isoform-level quantification from the transcription. | [40] |
| CircAST | https://github.com/xiaofengsong/CircAST]]> | A tool to assemble full-length circRNA transcripts and estimate their expression by using multiple splice graphs. | [41] |
| MapSplice | http://www.netlab.uky.edu/p/bioinfo/MapSplice]]> | A second algorithm for the alignment of RNA-seq reads to splice junctions. | [42] |
| circRNA_finder | https://github.com/orzechoj/circRNA_finder.git]]> | A de novo circRNA forecasting tool without the need of gene annotations. | [43] |
| segemehl | www.bioinf.uni-leipzig.de/Software/segemehl/]]> | A novel, unbiased algorithm to detect splice junctions from single-end cDNA sequences. | [44] |
| KNIFE | https://github.com/lindaszabo/KNIFE]]> | Using a statistical approach to discover and quantify circular and linear RNA splicing events. | [45] |
| DCC | https://github.com/dieterich-lab/]]> | Detection of back-splice junctions and circRNA versus host gene expression. | [46] |
| UROBORUS | http://uroborus.openbioinformatics.org/]]> | Detection of circRNAs with low expression levels in total RNA-seq. | [47] |
| NCLcomparator | https://github.com/TreesLab/NCLcomparator]]> | Non-co-linear (NCL, circular, intragenic trans-spliced or fusion RNAs) detecte tool. | [48] |
| CircMarker | https://github.com/lxwgcool/CircMarker]]> | Take advantage of transcription annotation files to create k-mer table for circular RNA detection. | [49] |
| CircDBG | https://github.com/lxwgcool/CircDBG]]> | A new method for circular RNA detection with De Bruijn graph. | [50] |
| find_circ | https://github.com/marvin-jens/find_circ]]> | The first RNA-Seq-based circRNA prediction tool. | [51] |
| CircExplorer2 | https://github.com/YangLab/CIRCexplorer2]]> | Annotating different types of alternative splicing events in circRNAs. | [52] |
| CIRI2 | https://sourceforge.net/projects/ciri/files/CIRI2]]> | Using an adapted maximum likelihood estimation to identify back splicing junction reads and to filter mapping errors. | [53] |
| CircSplice | https://github.com/GeneFeng/CircSplice]]> | Identify internal alternative splicing in circRNA and compare differential circRNA splicing events. | [54] |
| CirComPara | http://github.com/egaffo/CirComPara]]> | An bioinformatics pipeline to detect, quantify and annotate circRNAs from RNA-seq data. | [55] |
| CircRNAwrap | https://github.com/liaoscience/circRNAwrap]]> | Measuring the effectiveness of existing tools on collected and simulated data. | [56] |
| PcircRNA_finder | https://github.com/bioinplant/PcircRNA_finder/]]> | The frist circRNA prediction software for plants. | [57] |
| CircPlant | http://bis.zju.edu.cn/circplant]]> | With the incorporation of several plant-specific criteria detect plant circRNAs and predict function. | [58] |
| PCirc | https://github.com/Lilab -SNNU/Pcirc]]> | Using a machine learning method to predict plant circRNAs from RNA-seq data. | [59] |
随着大量植物circRNA被鉴定出来,一系列储存和可视化植物circRNA的数据库被建立(表 2,在刘旭庆等[60]基础上补充),这些数据库具有不同功能,不仅包含大量植物circRNA的序列、基因注释和功能预测等信息,同时也提供互作分析、可视化和miRNA靶点预测等工具。例如PlantCircBase[64]拥有超过115 000个来自16种不同植物的circRNA,还提供了特定circRNA结构的可视化、相应物种中涉及circRNA-miRNA-mRNA的潜在相互作用网络、Sanger测序的验证信息以及功能注释、组织表达、保守性验证等更多信息,PlantCircBase有望成为研究植物circRNA最全面的数据库资源;GreenCircRNA[68]是第一个预测植物circRNA可以作为miRNA诱饵的数据库,收录了69种植物的21万多条circRNA,预测了38种植物的circRNA作为miRNA诱饵的可能性。这些数据库的建立为阐明植物circRNA的各种机制方面提供了巨大的帮助。
| Tool names | Uniform resource locator | Description | References |
| AtCircDB | http://genome.sdau.edu.cn/circRNA]]> | A comprehensive tissue-specific database to help store, retrieve, visualize and download Arabidopsis circular RNAs. | [62] |
| PlantCircNet | http://bis.zju.edu.cn/plantcircnet/index.php]]> | An integrated database that provides visualized plant circRNA miRNA mRNA regulatory networks containing identified circRNAs in eight model plants. | [63] |
| PlantcircBase | http://ibi.zju.edu.cn/plantcircbase/]]> | Providing the most comprehensive information about plant circRNAs. | [64] |
| CircFunBase | http://bis.zju.edu.cn/CircFunBase]]> | A web-accessible functionally annotated circRNA database. | [65] |
| CropCircDB | http://deepbiology.cn/crop/]]> | A comprehensive collection of circRNAs in crop response to abiotic stress. | [66] |
| ASmiR | http://forestry.fafu.edu.cn/bioinfor/db/ASmiR]]> | A database of miRNA targets in alternatively spliced linear and circRNAs for plant. | [67] |
| GreenCircRNA | http://greencirc.cn]]> | The first database for the prediction of plant circRNAs that act as miRNA decoys. | [68] |
2012年,Salzman等首次发现pre-mRNA通过反向剪接将3′下游剪接供体连接到5′上游剪接受体上形成circRNA[28],催化pre-mRNA剪接的U2依赖型剪接体(U2-dependent spliceosome) 剪接5′端供体位点“GU”和3′端受体位点“AG”信号[69]。已有研究证明典型的剪接机制和剪接体信号都是反向剪接环化所必需的[70-71]。大多数高表达circRNA通常由pre-mRNA的内部外显子加工而成,包含多个外显子,说明反向剪接通常与典型剪接结合[72]。在拟南芥和水稻的研究中发现分别有13%和34%的circRNA通过反向剪接形成[29]。拟南芥中99%的circRNA均具有典型的GU/AG剪接信号[73],而在水稻circRNA中仅有7.3%具有典型GU/AG剪接信号,其余为不同的非典型剪接信号[39],此外葡萄[22]、棉花(Gossypium spp.)[74]等植物circRNA形成过程中也依赖非典型剪接信号,表明植物circRNA中存在特定的剪接信号模式,但是还需要在更多的植物中进一步验证。
3.2 表达特异性circRNA在植物中通常表现出不同组织、细胞和发育阶段的特定表达模式,既有表达有无的差异,也有表达量的差异。其在拟南芥和水稻的根、茎、叶和种子等组织中均有表达,但是表达量不同,且有很大差异[6, 29];大豆和猕猴桃根、茎、叶组织中具有不同表达量的circRNA,而且在不同组织中特异性表达[18, 21]。在拟南芥中叶绿体circRNA数量多于线粒体,在不同细胞中特异性表达,暗示circRNA可能参与植物光合作用和呼吸作用[73]。在早花枳壳(Poncirus trifoliata L. Raf.) 突变体及其野生型转录组数据分析中发现,176个差异表达的circRNA可能在早花过程中起重要作用[75];circRNA circbHLH93在毛竹(Phyllostachys edulis (Carr.) Mitford cv. Pubescens) 8个不同发育阶段的竹笋中存在表达差异[76];来源于八氢番茄红素合成酶1 (PSY1)和八氢番茄红素脱氢酶(PDS)的circRNA被发现在番茄果实成熟的不同阶段有不同的表达[77];在水稻从正常发育到衰老的剑叶鉴定到的6 612个circRNA中发现其中113个circRNA在叶片衰老过程中有差异表达[78]。同时,植物circRNA也在不同逆境下特异性表达,例如在水稻磷酸盐失衡[29]、小麦干旱[19]和葡萄低温[22]等不同胁迫条件下circRNA的表达量均发生差异变化。
3.3 保守性各种RNA-seq数据显示了植物不同物种circRNA的保守性质。水稻和拟南芥中有超过700个circRNA的来源基因存在同源性,并有300多个同源基因从相似的位置产生circRNA[29];对拟南芥、水稻和大豆可以产生circRNA的8 362、9 385和1 995个来源基因进行保守性分析发现,大豆和拟南芥之间有685个同源,大豆与水稻之间有1 095个同源,3个物种中共有551个来源基因同源[18]。杨树(Populus tomentosa Carr.) 响应干旱的circRNA来源基因中有一半与拟南芥和玉米同源,表明参与干旱响应转录调控的circRNA在单子叶和双子叶植物中是保守的[79]。CircRNA的序列保守性表明,这些circRNA可能具有相似的潜在生物学功能,这些保守的circRNA在植物中的功能还需要进一步的研究和验证。
3.4 稳定性与线性RNA相比,两端共价闭合和二级结构的存在使circRNA更稳定,具有核糖核酸外切酶(如RNase R) 抗性,因此不易降解[80]。RNase R能降解线性RNA但不能降解circRNA,用其可以提高检测circRNA的灵敏度,减少假阳性的数量。Zeng等[75]通过实时荧光定量PCR (real-time PCR) 实验对11个已证实的枳壳circRNA进行了抗RNase R测验,证实了植物circRNA的RNase R抗性。且circRNA半衰期的中位数超过48 h,而它对应的线性RNA半衰期的中位数不到20 h,证明了circRNA在细胞内具有高度稳定性[81]。
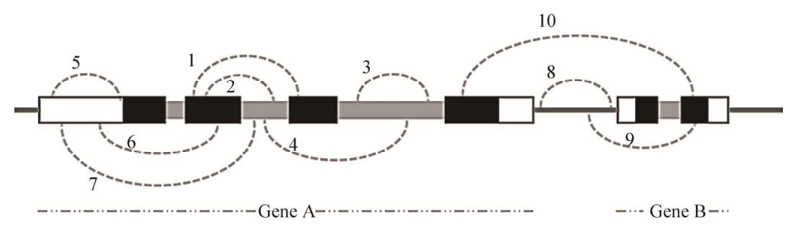
4 植物circRNA的类型随着研究的深入,circRNA的类型不断积累,是研究的重点关注领域之一[82],主要的3种类型分别是外显子circRNA、内含子circRNA和外显子-内含子circRNA[83]。Chu等[25]根据circRNA在基因组中两个剪接位点的位置,将circRNA分为10种类型(图 1,在Chu等[25]基础上修改)。此外,还有一些其他的circRNA,例如融合基因来源的circRNA (fusion-circRNA, f-circRNA)[84-85]、聚合酶Ⅱ转录通读形成的通读circRNA (read-through circRNA, rt-circRNA)[86]、具有重叠区域和不同位点的circRNA,称作相互包容的circRNA (mutually inclusive circular RNAs)[76]等,不同的研究方法和分类方式以及可变剪接的存在使我们更大程度上认识到了circRNA的多样性,因此能开展更深入地研究。
5 植物circRNA的生成机制CircRNA来源于pre-mRNA,由RNA聚合酶Ⅱ(Pol Ⅱ) 转录[70-72]。CircRNA的生成调控依赖于控制剪接的顺式调控元件和反式作用因子。除此之外,通过核酶自剪切也可以调控植物circRNA的生成。
|
| 图 1 circRNA的类型[25] Fig. 1 Types of circRNAs[25]. 1: e-circRNA, two back-splicing sites of a circRNA are both at exons; 2: ei-circRNA, one back-splicing site of a circRNA is at exon while the other is at intron; 3: i-circRNA, two back-splicing sites of a circRNA are both at a single intron; 4: ie-circRNA, two back-splicing sites of a circRNA are at two different introns across one or several exons; 5: u-circRNA, two back-splicing sites of a circRNA are both at UTRs; 6: ue-circRNA, one back-splicing site of a circRNA is at UTR while the other is at exon; 7: ui-circRNA, one back-splicing site of a circRNA is at UTR while the other is at intron; 8: ig-circRNA, two back-splicing sites of a circRNA are both at a single intergenic region; 9: igg-circRNA, one back-splicing site of a circRNA is at intergenic region while the other is at genic region; 10: ag-circRNA, two back-splicing sites of a circRNA are at two different genes. The black, gray and blank bars represent exons, introns and UTRs, respectively. The black lines represent intergenic region of the genomes. |
| |
研究发现侧翼内含子序列互补有助于大多数外显子环化,Gao等通过烟草瞬时表达葡萄circRNA发现侧翼内含子的长度会影响成环效率,且长度越长,成环效率越高,短至40 nt的侧翼内含子重复序列足以实现外显子环化[22, 87-88]。在动物环化外显子的侧翼内含子中富集反向互补序列(reverse complementary pairs, RCPs),通常来源于重复元件,例如RCMs (reverse complementary matches)[88]、ICSs (intronic complementary sequences)[89]及Alu元件[88]。然而,植物外显子circRNA的侧翼内含子中具有有限的RCPs,例如,在水稻、拟南芥和大豆中,RCPs的比例分别为6.2%、2.7%和0.3%[6, 18, 29],在13个经过验证的水稻circRNA中,只有两个在其侧翼内含子中包含 > 15 bp的RCPs[29],另一项研究在水稻外显子circRNA侧翼内含子序列中发现微型反向重复转座元件(miniature inverted repeat transposable elements, MITEs)[6];在毛竹circRNA侧翼内含子中富集长终端重复转座元件(long terminal repeat transposable elements, LTRTEs)[90];在玉米circRNA侧翼区域发现LINE1-like元件(long interspersed nuclear element 1-like elements, LLEs)及其RCPs (LLERCPs)[20],随着LLERCPs数量增加,circRNA积累量上升;在杨树circRNA侧翼内含子区域发现的完整MITE元件及其RCPs,对杨树Circ_0003418的生成极为重要[79],证明了转座元件对植物circRNA的生成可能起到重要作用,为内含子互补促进环化的机制提供了充足的证据。
5.1.2 外显子跳读当pre-mRNA进行经典的GU/AG剪接时,可以发生跨外显子的剪接方式,即外显子跳读,产生包含内含子-外显子的套索中间体,随后该中间体发生反向剪接形成circRNA[91]。内含子circRNA是套索驱动模型中的特殊方式,其依赖于5′剪接位点的7 nt富含GU基序和靠近分支位点的11 nt富含C基序,通过聚合酶Ⅱ的转录形成一个套索内含子,最终通过2′-5′磷酸二酯键共价连接而环化,接着从内含子3′端到分支位点的多余序列被降解[31]。目前在拟南芥、番茄、水稻和玉米中的研究表明,这种套索结构在植物中是广泛存在的,并识别到了大量由套索结构驱动环化的circRNA[92]。在拟南芥中,SEPALLATA3 (SEP3) 的第六个外显子跳读生成的circRNA与宿主基因DNA杂交形成RNA: DNA-环,从而延缓转录延伸,以增强外显子跳读引起的环状转录本生成,此研究为circRNA生成与外显子跳读之间的关系提供了证据[93]。
5.2 反式作用因子调控除了顺式元件外,反式作用因子,如RNA结合蛋白(RNA binding protein, RBP) 在某些条件下,也可作为circRNA生成的激活剂或抑制剂,如musclebling (MBL)[70]、quaking (QKI)[94-95]、adenosine deaminase 1 (ADAR1)[96]、fused in sarcoma (FUS)[97]和DEAH-box helicase 9 (DHX9)[98]等,虽然已在拟南芥中鉴定出这些RBP的同源蛋白,但它们在circRNA生成过程中的潜在功能仍有待研究。在人类上皮-间质细胞转换中,QKI通过结合到宿主RNA侧翼内含子上形成二聚体,促进内含子-内含子之间相互作用使3′端和5′端接近以进行环化[94-95]。拟南芥中的QKI同源蛋白包含26个KH结构域,其中有5个与QKI高度相似,被证明参与pre-mRNA加工[99],在植物的开花调节[100-101]、应激反应[102-104]和激素信号转导[105]中起重要作用,同时也可能与QKI蛋白具有相似功能,例如参与circRNA生成[106],但这只是一个推测,并未有更多报道来证实或推翻。大多数RBP促进circRNA生成,但也有部分抑制环化。例如DHX9是一个有RNA结合功能域和解旋酶功能域的核RNA解旋酶,通过与反向互补Alu元件相结合,之后行使RNA解旋酶功能,导致Alu元件的解旋进而抑制circRNA的形成[98]。目前关于植物RBP调控促进或抑制circRNA生成的报道仍很少。
5.3 核酶自剪切植物circRNA还可能通过核酶自剪切来协助产生,最早报道的circRNA含有小的自切割RNA基序,如锤头状核酶(hammerhead ribozymes, HHR) 和发夹核酶(hairpin ribozymes, HR)[7, 107],HDV也编码自切割基序,称为HDV核酶[108-109]。这些核酶的参与会有效促进circRNA的积累。例如对几种植物如麻风树(Jatropha curcas L.)、草莓(Fragaria × ananassa Duch.)、桉树(Eucalyptus robusta Smith)或柑橘(Citrus L.) 的不同体细胞和生殖组织进行Northern blotting分析和RT-PCR实验表明,植物基因组中存在两侧串联有Ⅲ型HHR基序并能精确表达为环状RNA的序列[110]。另一项研究预测了夏威夷群岛无花果树(Ficus carica Linn.) 相关的类病毒RNA,其中一个RNA被鉴定为大小在357–360个核苷酸之间的circRNA,这个类病毒circRNA在每条极性链上都含有HHR基序,在转录过程中,HHR被证明在预测的位点上进行自切割[111]。除反向剪接外,通过HHR的参与来促进circRNA的表达被认为是第二种circRNA生成的途径,具有作为基因调控新形式的潜力[112]。
6 植物circRNA的功能由于circRNA的低水平存在导致人们的研究受到极大阻碍,到目前为止,大多数circRNA功能不够明确。最近的研究发现,植物circRNA可以作为miRNA海绵,通过竞争性内源RNA机制来调节基因表达,同时也具有蛋白质编码潜力,此外植物circRNA在激素刺激和逆境胁迫等条件下也呈现差异表达模式,影响植物生长发育和参与细胞间信号传递,初步体现了其功能的多样性,相信在未来植物circRNA在功能发挥和应用潜力上的研究会越来越深入和多样化。
6.1 作为miRNA海绵circRNA已证实的功能之一是通过发挥miRNA海绵吸附作用直接或间接结合靶标miRNA,阻断miRNA对其靶基因的调控[113]、建立circRNA-miRNA-mRNA网络和影响基因表达和转录调控。目前鉴定到的不同植物circRNA中都预测到一定比例的circRNA是潜在的miRNA靶标,拟南芥中有5.0%的circRNA具有作为miRNA海绵的潜在能力[29];水稻1 356个外显子circRNA中发现235个推测的miRNA结合位点,只有31个circRNA含有两个或两个以上的miRNA结合位点[6];玉米2 804个circRNA中有15个miRNA结合位点[20];枳壳29个circRNA可以作为16个miRNA的潜在靶标[75];circRNA45和circRNA47可能作为miR477-3P海绵来调节靶标抗性基因SpRLK1/2的表达,在番茄对晚疫病的免疫中起到正调节作用[114];另一项研究比较野生型和乙烯信号转录因子(ethylene-responsive transcription factors, LeERF1) 转基因番茄果实发现61个circRNA可能具有吸附miRNA的作用,其中一些miRNA已被发现参与乙烯信号转导途径[115];在棉花差异表达的circRNA中发现,其中有7个可以与17个miRNA相结合参与棉花纤维发育的调控[116];在水稻中,circRNA Os08Circ16564被预测为miR172的目标模拟物,但是RT-PCR结果表明,在Os08Circ16564过表达植株中miR172的表达水平与野生型没有显著差异,表明Os08circ16564可能不是miR172的真正海绵[6]。一个miRNA可以靶向多个circRNA,同时一个circRNA也可以被几个不同的miRNA作为靶标,但是到目前为止,在各种生物体中发现的大多数circRNA表达量较低,很少包含相同miRNA的多个结合位点,因此许多circRNA似乎不可能作为miRNA海绵发挥作用。此外即使预测到miRNA结合位点,也仅停留在预测阶段或者出现预测错误的情况,因此需要使用更多候选circRNA来进行实验验证。
6.2 具有翻译潜力circRNA被归入非编码RNA就是由于其翻译潜力未得到太多关注。近年来有研究证明了circRNA的蛋白质编码潜力[117-119]。例如存在于果蝇头部的circMbl,可以产生37.04 kDa的蛋白质[118];人类Circ-ZNF609包含一个完整的开放读码阅读框(open reading frame, ORF),具有起始和终止密码子,可以被内部核糖体进入位点(internal ribosome entry site, IRES) 翻译成蛋白质[117];除IRES外,在存在修饰位点的情况下翻译带有ORF的circRNA,翻译效率与N6-甲基腺苷(N6-methylation of adenosine, m6A)位点的数量呈正相关[119]。目前,在大豆响应低温胁迫的3个circRNA中预测到其至少包含一个IRES元件和一个ORF[120];Han等在玉米中预测到229个circRNA具有编码潜力,发现较长的circRNA具有更高的编码潜力[121];在拟南芥中发现m6A的修饰位点通常位于mRNA的起始和终止密码子附近[122-123],暗示植物circRNA也具有翻译潜力。
6.3 响应生物/非生物胁迫多项研究表明,circRNA响应生物/非生物胁迫,在植物发育过程中发挥重要作用。Ye等发现水稻中有27个外显子circRNA在磷酸盐充足和饥饿条件下差异表达[29];番茄和拟南芥中分别有163个和1 583个circRNA在冷热处理条件下差异表达[17, 124];小麦有62个circRNA在脱水胁迫条件下的幼苗中有差异表达[19];冷胁迫处理不同时间段,共有475个葡萄circRNA差异表达[22];黄瓜(Cucumis sativus Linn.) circRNA在盐胁迫中差异表达[125];在拟南芥中过表达来源于外向整流型钾离子通道基因circGORK,发现转基因植株的种子萌发对脱落酸(ABA) 超敏感,且抗旱性增强,为circRNA直接调控干旱胁迫提供了有力证据[126]。
除了非生物胁迫,circRNA也被报道对生物胁迫有反应。例如,大豆中199个circRNA在棉铃虫取食叶片损伤胁迫下的抗病和感病样本之间存在差异表达[127];猕猴桃中584个差异表达的circRNA响应丁香假单胞猕猴桃致病变种(Pseudomonas syringae pv. actinidiae, PSA) 的感染[21];马铃薯(Solanum tuberosum L.) 中429个差异表达的circRNA响应胡萝卜软腐果胶杆菌巴西亚种(Pectobacterium carotovorum subsp. brasiliense, PCB) 的感染[128],在PCB感染时,circRNA在感病品种中表达下调,抗病品种中表达上调;棉花感染黄萎病菌(Verticillium) 后共发现280个差异表达的circRNA,且易感病株系中差异表达的circRNA数约为抗病株系的两倍[129];番茄感染黄化曲叶病毒(tomato yellow leaf curl virus, TYLCV) 病叶与对照之间分别有32个和83个circRNA特异表达,且感染病毒后circRNA的表达量低于对照[130];过量表达CircR5g05160的转基因水稻可提高对稻瘟病菌(Magnaporthe oryzae) 的抗病性[131]。这些结果表明,circRNA可能在植物对生物/非生物胁迫的应答中发挥着重要而多样的功能,另外这些差异表达的circRNA也可能作为植物中有效的生物标记。
除此之外,植物circRNA还有其他功能,例如过表达拟南芥AT5G37720的第一个内含子产生的套索circRNA (lariat41)发现能调节基因表达并影响拟南芥发育[132],过表达植株(lariat41-OE) 表现出叶片卷曲和簇生、开花晚和育性低的特点,并伴随800多个基因表达量的改变[133]。其中关键的开花时间调节因子FT在lariat41-OE植株中的表达水平显著降低,可能是导致出现晚花表型的原因。此项研究表明,circRNA通过调控基因的表达量从而影响植株的生长发育,但只是少数circRNA在功能上得到了验证,具体如何调控还需进一步了解。植物circRNA还可能作为信号分子参与细胞之间信号传递和木质部韧皮部的长距离运输。例如马铃薯纺锤块茎类病毒(potato spindle tuber viroid, PSTVd) 通过韧皮部的长距离运输强调了circRNA通过植物维管系统传递分子的可能性[134-135],推测植物内源circRNA可能采取类似的方式参与信号传递,因此在这个方向上需要更广泛地研究。植物circRNA的功能在很大程度上仍未被探索,但越来越多的证据开始显示,植物circRNA在许多生物过程中发挥着重要作用。
7 circRNA的降解与定位circRNA没有自由末端,因此并不能沿用大多数RNA降解途径。MiRNA介导的circRNA降解是目前得到比较明确证明的circRNA降解途径,例如circRNA CDR1as被miR-671介导的核酸内切酶(argonaute-2, Ago2) 切割[136]。miR-671在CDR1as的结合位点与miRNA完全互补,导致Ago2切割[113],但是其他circRNA是否能通过miRNA介导的途径降解尚不清楚。m6A可促进潜在可降解circRNA的核酸内切酶的招募[137],其修饰的circRNA被人类YTH结构域家族2 (YTH domain containing family 2, YTHDF2) 蛋白识别,结合的环状转录本被选择性降解[138],另一项研究发现HeLa细胞经poly (I: C) 刺激或病毒感染即会激活核酸内切酶RNase L,从而导致整体circRNA的降解[139]。此外,果蝇DL1/S2细胞中GW182缺失后,circRNA水平升高[140],GW182通常被认为是miRNA介导的基因沉默关键因子,且其同源物在调节HeLa细胞circRNA降解方面显示出类似的作用[141],因此表明GW182对circRNA的降解起特异性作用[142]。Guria等通过计算预测了大约85%的miRNA与植物circRNA的完美互补结合位点[143],这些位点可能使circRNA作为miRNA海绵导致miRNA活性被抑制,也可能是miRNA在植物中切割circRNA的一种调控机制,miRNA中第10和第11核苷酸的序列可能决定了circRNA是作为miRNA的海绵,还是被miRNA切割。如果miRNA中的第10和第11个核苷酸与circRNA完全匹配,miRNA可能会有更多的机会切割和降解circRNA[144]。目前我们对于植物circRNA的降解仍知之甚少,因此需要更多的研究来探索。
一些报告证实细胞质中存在外显子circRNA[50, 80, 145],而保留内含子的circRNA,如外显子-内含子circRNA和内含子circRNA,仅在细胞核中发现[31, 146-147]。但是,在Neuro2a (N2a) 细胞中,发现外显子circRNA也定位在细胞核中[98],表明外显子circRNA的定位仍不清楚。此外发现,circRNA一旦在细胞核中产生,通常被运输到细胞质中,以实现其正常的功能或降解[28, 148]。Sun等发现circRNA中存在多个内部互补碱基配对序列(internal complementary base-pairing sequences, ICBPS),推测其可能不是简单的环状结构,还含有双链结构,这种结构的形成有助于circRNA与RBP牢固的结合,从而便于它们从细胞核输出到细胞质中,并且双链结构可能使circRNA更容易被相关的酶降解[149]。
8 讨论与展望随着测序技术和生物信息学方法的发展,越来越多的circRNA被鉴定出来,然而使用不同算法在同一物种中鉴定的circRNA的数量不同,假阳性占比也很高,因此需要不同测序技术、测序深度、预测工具和算法以及处理方法等同时结合来提高circRNA鉴定的准确性和精度。同时得益于测序技术的不断改进,circRNA的神秘面纱逐渐被揭开。目前检测circRNA常用的二代测序根据短片段进行RNA组装获得全长circRNA,易出现线性转录本污染问题且很难准确定量,三代测序的出现解决了这个问题,它能够实现 > 10 kb长度circRNA的全序列组装,使人们真正看到circRNA的完整序列,可识别circRNA隐藏的复杂结构,并且其不需要扩增,实现了对基因组的均匀覆盖,能检测到更多低水平表达和新的circRNA,准确度、检出率和灵敏度大大提升[150-152]。大规模获得全长circRNA有助于人们理解其内部结构及可变剪切方式,进而揭示更多circRNA的功能和调控过程。
在鉴定出大量circRNA的同时人们对于circRNA的认识也达到了新的层面,circRNA的生成机制和功能应用已成为一个相关的研究课题。在动物中,circRNA两侧内含子的长度明显大于所有内含子的平均长度,这与在植物中的发现一致[29],这些侧翼内含子还包含介导circRNA生成的RCPs[153]。植物侧翼内含子含有与动物相比较少的重复序列,表明植物与动物circRNA的生成机制可能不同。植物侧翼内含子中包含的转座子元件和RCPs对circRNA生成的影响还有待进一步研究。同时植物和动物之间circRNA生成的差异也将是一个有趣的研究方向。
在circRNA的功能方面,circRNA在生物体中动态表达,与mRNA、miRNA及lncRNA等形成互作网络,参与调控宿主基因的转录及转录后表达。植物circRNA目前的研究主要集中在预测及鉴定工作,对于它们的功能认知还不是很明朗,由于circRNA来源于与线性RNA相同的pre-RNA[80],所以很难独立于线性RNA来研究circRNA,这使得用传统的RNAi很难沉默circRNA[154]。随着CRISPR-Cas技术的发展[155-158],为研究circRNA的功能提供了新的思路,其在分析非编码基因功能方面特别强大,可以通过靶向缺失产生空等位基因,这是传统诱变工具难以实现的。Zhou等基于CRISPR-Cas9介导的基因组长片段缺失策略实现了4个水稻circRNA的敲除与功能鉴定。该研究不仅提供了植物circRNA位点通过海绵吸附响应来负调控miRNA的明显证据,而且还为进一步通过编辑植物circRNA位点改良农艺性状提供了新思路[158]。杨树MITE元件及其反向互补序列的存在为利用CRISPR-Cas9进行杨树遗传改良提供了理想的大片段缺失靶位点[79]。最近出现的CRISPR-Cas13允许通过靶向跨越反向剪接接头的序列来识别circRNA,有效而特异地区分了circRNA和mRNA,同时能够高效地降低circRNA表达而不影响其来源线性mRNA表达,使CRISPR-Cas13成为在单个和大规模水平上发现和研究circRNA功能的有用工具[157, 159-160]。以circRNA为靶点的基因编辑在植物circRNA功能探究中的应用正处于新兴阶段,是研究的热点之一。
此外,circRNA在不同植物物种中的鉴定尚不全面,如柑橘、油菜、大麦和小麦等的circRNA鉴定数量仍然较少。对于植物circRNA在细胞中的定位、输出和降解机制等尚不清楚。此外植物circRNA特异性环化的原因、以何种机制启动环化、环化后是否影响来源基因及临近基因表达,会产生什么样的生物学效应等具体问题仍未可知,解决好具体问题将有助于研究者开展未来的探索,同时也有助于拓宽植物circRNA的研究领域。虽然在过去几年植物circRNA的研究进展迅猛,但是同动物相比,仍处于起步阶段,随着人们的广泛关注,相信在将来会对植物circRNA有一个全面的认知。
| [1] |
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell, 2009, 136(4): 629-641. DOI:10.1016/j.cell.2009.02.006
|
| [2] |
Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature, 2012, 489(7414): 101-108. DOI:10.1038/nature11233
|
| [3] |
Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol, 2013, 64: 137-159. DOI:10.1146/annurev-arplant-050312-120043
|
| [4] |
Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet, 2019, 20(11): 675-691. DOI:10.1038/s41576-019-0158-7
|
| [5] |
Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One, 2014, 9(6): e90859.
|
| [6] |
Lu T, Cui L, Zhou Y, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA, 2015, 21(12): 2076-2087. DOI:10.1261/rna.052282.115
|
| [7] |
Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. PNAS, 1976, 73(11): 3852-3856. DOI:10.1073/pnas.73.11.3852
|
| [8] |
Arnberg AC, Van Ommen GJB, Grivell LA, et al. Some yeast mitochondrial RNAs are circular. Cell, 1980, 19(2): 313-319. DOI:10.1016/0092-8674(80)90505-X
|
| [9] |
Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature, 1986, 323(6088): 558-560. DOI:10.1038/323558a0
|
| [10] |
Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. PNAS, 1990, 87(19): 7628-7632. DOI:10.1073/pnas.87.19.7628
|
| [11] |
Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell, 1991, 64(3): 607-613. DOI:10.1016/0092-8674(91)90244-S
|
| [12] |
Fan X, Zhang X, Wu X, et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol, 2015, 16: 148. DOI:10.1186/s13059-015-0706-1
|
| [13] |
Shen Y, Guo X, Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett, 2017, 591(1): 213-220. DOI:10.1002/1873-3468.12500
|
| [14] |
Cortes-Lopez M, Gruner MR, Cooper DA, et al. Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genom, 2018, 19(1): 8. DOI:10.1186/s12864-017-4386-y
|
| [15] |
Danan M, Schwartz S, Edelheit S, et al. Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res, 2012, 40(7): 3131-3142. DOI:10.1093/nar/gkr1009
|
| [16] |
Darbani B, Noeparvar S, Borg S. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front Plant Sci, 2016, 7: 776.
|
| [17] |
Zuo J, Wang Q, Zhu B, et al. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun, 2016, 479(2): 132-138. DOI:10.1016/j.bbrc.2016.07.032
|
| [18] |
Zhao W, Cheng Y, Zhang C, et al. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep, 2017, 7(1): 5636. DOI:10.1038/s41598-017-05922-9
|
| [19] |
Wang Y, Yang M, Wei S, et al. Identification of circular RNAs and their targets in leaves of Triticum aestivum L.under dehydration stress. Front Plant Sci, 2016, 7: 2024.
|
| [20] |
Chen L, Zhang P, Fan Y, et al. Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol, 2018, 217(3): 1292-1306. DOI:10.1111/nph.14901
|
| [21] |
Wang Z, Liu Y, Li D, et al. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci, 2017, 8: 413.
|
| [22] |
Gao Z, Li J, Luo M, et al. Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol, 2019, 180(2): 966-985. DOI:10.1104/pp.18.01331
|
| [23] |
Wang J, Lin J, Wang H, et al. Identification and characterization of circRNAs in Pyrus betulifolia Bunge under drought stress. PLoS One, 2018, 13(7): e0200692. DOI:10.1371/journal.pone.0200692
|
| [24] |
Dong Y, Chen H, Gao J, et al. Bioactive ingredients in Chinese herbal medicines that target non-coding RNAs: promising new choices for disease treatment. Front Pharmacol, 2019, 10: 515. DOI:10.3389/fphar.2019.00515
|
| [25] |
Chu QJ, Bai PP, Zhu XT, et al. Characteristics of plant circular RNAs. Brief Bioinform, 2018, 21(1): 135-143.
|
| [26] |
Chu Q, Shen E, Ye C Y, et al. Emerging roles of plant circular RNAs. Plant Cell Dev, 2018, 1(1): 1-14.
|
| [27] |
Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. Faseb J, 1993, 7(1): 155-160. DOI:10.1096/fasebj.7.1.7678559
|
| [28] |
Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One, 2012, 7(2): e30733. DOI:10.1371/journal.pone.0030733
|
| [29] |
Ye CY, Chen L, Liu C, et al. Widespread noncoding circular RNAs in plants. New Phytol, 2015, 208(1): 88-95. DOI:10.1111/nph.13585
|
| [30] |
Meng X, Chen Q, Zhang P, et al. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics, 2017, 33(20): 3314-3316. DOI:10.1093/bioinformatics/btx446
|
| [31] |
Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell, 2013, 51(6): 792-806. DOI:10.1016/j.molcel.2013.08.017
|
| [32] |
You X, Conrad TO. Acfs: accurate circRNA identification and quantification from RNA-Seq data. Sci Rep, 2016, 6: 38820. DOI:10.1038/srep38820
|
| [33] |
Jia GY, Wang DL, Xue MZ, et al. CircRNAFisher: a systematic computational approach for de novo circular RNA identification. Acta Pharmacol Sin, 2019, 40(1): 55-63. DOI:10.1038/s41401-018-0063-1
|
| [34] |
Jakobi T, Uvarovskii A, Dieterich C. Circtools-a one-stop software solution for circular RNA research. Bioinformatics, 2019, 35(13): 2326-2328. DOI:10.1093/bioinformatics/bty948
|
| [35] |
Metge F, Czaja-Hasse LF, Reinhardt R, et al. FUCHS-towards full circular RNA characterization using RNAseq. Peer J, 2017, 5: e2934. DOI:10.7717/peerj.2934
|
| [36] |
Gao Y, Wang H, Zhang H, et al. PRAPI: post-transcriptional regulation analysis pipeline for Iso-Seq. Bioinformatics, 2018, 34(9): 1580-1582. DOI:10.1093/bioinformatics/btx830
|
| [37] |
Feng J, Xiang Y, Xia S, et al. CircView: a visualization and exploration tool for circular RNAs. Brief Bioinform, 2018, 19(6): 1310-1316.
|
| [38] |
Zhong S, Wang J, Zhang Q, et al. CircPrimer: a software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinformatics, 2018, 19(1): 292. DOI:10.1186/s12859-018-2304-1
|
| [39] |
Ye CY, Zhang X, Chu Q, et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol, 2017, 14(8): 1055-1063. DOI:10.1080/15476286.2016.1245268
|
| [40] |
Zheng Y, Ji P, Chen S, et al. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med, 2019, 11(1): 2. DOI:10.1186/s13073-019-0614-1
|
| [41] |
Wu J, Li Y, Wang C, et al. CircAST: full-length assembly and quantification of alternatively spliced isoforms in circular RNAs. Genomics Proteomics Bioinformatics, 2019, 17(5): 522-534. DOI:10.1016/j.gpb.2019.03.004
|
| [42] |
Wang K, Singh D, Zeng Z, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res, 2010, 38(18): e178. DOI:10.1093/nar/gkq622
|
| [43] |
Westholm JO, Miura P, Olson S, et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep, 2014, 9(5): 1966-1980. DOI:10.1016/j.celrep.2014.10.062
|
| [44] |
Hoffmann S, Otto C, Doose G, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol, 2014, 15(2): R34. DOI:10.1186/gb-2014-15-2-r34
|
| [45] |
Szabo L, Morey R, Palpant NJ, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol, 2015, 16: 126. DOI:10.1186/s13059-015-0690-5
|
| [46] |
Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics, 2016, 32(7): 1094-1096. DOI:10.1093/bioinformatics/btv656
|
| [47] |
Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res, 2016, 44(9): e87. DOI:10.1093/nar/gkw075
|
| [48] |
Chen CY, Chuang TJ. NCLcomparator: systematically post-screening non-co-linear transcripts (circular, trans-spliced, or fusion RNAs) identified from various detectors. BMC Bioinform, 2019, 20(1): 3. DOI:10.1186/s12859-018-2589-0
|
| [49] |
Li X, Chu C, Pei J, et al. CircMarker: a fast and accurate algorithm for circular RNA detection. BMC Genomics, 2018, 19(suppl 6): 572.
|
| [50] |
Li X, Wu YF. Detecting circular RNA from high-throughput sequence data with De Bruijn graph. BMC Genom, 2020, 21(Suppl 1): 749.
|
| [51] |
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 2013, 495(7441): 333-338. DOI:10.1038/nature11928
|
| [52] |
Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res, 2016, 26(9): 1277-1287. DOI:10.1101/gr.202895.115
|
| [53] |
Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform, 2018, 19(5): 803-810. DOI:10.1093/bib/bbx014
|
| [54] |
Feng J, Chen K, Dong X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer, 2019, 18(1): 35. DOI:10.1186/s12943-019-0996-0
|
| [55] |
Gaffo E, Bonizzato A, Kronnie GT, et al. CirComPara: a multi-method comparative bioinformatics pipeline to detect and study circRNAs from RNA-seq data. Noncoding RNA, 2017, 3(1): E8.
|
| [56] |
Li L, Bu D, Zhao Y. CircRNAwrap-a flexible pipeline for circRNA identification, transcript prediction, and abundance estimation. FEBS Lett, 2019, 593(11): 1179-1189.
|
| [57] |
Chen L, Yu Y, Zhang X, et al. PcircRNA_finder: a software for circRNA prediction in plants. Bioinformatics, 2016, 32(22): 3528-3529.
|
| [58] |
Zhang P, Liu Y, Chen H, et al. CircPlant: an integrated tool for circRNA detection and functional prediction in plants. Genomics Proteomics Bioinformatics, 2020, 18(3): 352-358. DOI:10.1016/j.gpb.2020.10.001
|
| [59] |
Yin S, Tian X, Zhang J, et al. PCirc: random forest-based plant circRNA identification software. BMC Bioinformatics, 2021, 22(1): 10. DOI:10.1186/s12859-020-03944-1
|
| [60] |
刘旭庆, 高宇帮, 赵良真, 等. 环状RNA的产生、研究方法及功能. 遗传, 2019, 41(6): 469-485. Liu XQ, Gao YB, Zhao LZ, et al. Biogenesis, research methods, and functions of circular RNAs. Hereditas, 2019, 41(6): 469-485 (in Chinese). |
| [61] |
Chen L, Wang C, Sun H, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform, 2021, 22(2): 1706-1728. DOI:10.1093/bib/bbaa001
|
| [62] |
Ye J, Wang L, Li S, et al. AtCircDB: a tissue-specific database for Arabidopsis circular RNAs. Brief Bioinform, 2019, 20(1): 58-65. DOI:10.1093/bib/bbx089
|
| [63] |
Zhang PJ, Meng XW, Chen HJ, et al. PlantCircNet: a database for plant circRNA-miRNA-mRNA regulatory networks. Database (Oxford), 2017, 2017(10.1093): database.
|
| [64] |
Chu QJ, Zhang XC, Zhu XT, et al. PlantcircBase: a database for plant circular RNAs. Mol Plant, 2017, 10(8): 1126-1128. DOI:10.1016/j.molp.2017.03.003
|
| [65] |
Meng XW, Hu DH, Zhang PJ, et al. CircFunBase: a database for functional circular RNAs. Database (Oxford), 2019, 2019(10.1093): database.
|
| [66] |
Wang K, Wang C, Guo BH, et al. CropCircDB: a comprehensive circular RNA resource for crops in response to abiotic stress. Database (Oxford), 2019, 2019(10.1093): database.
|
| [67] |
Wang H, Wang H, Zhang H, et al. The interplay between microRNA and alternative splicing of linear and circular RNAs in eleven plant species. Bioinformatics, 2019, 35(17): 3119-3126. DOI:10.1093/bioinformatics/btz038
|
| [68] |
Zhang JJ, Hao ZQ, Yin SW, et al. GreenCircRNA: a database for plant circRNAs that act as miRNA decoys. Database (Oxford), 2020, 2020(10.1093): database.
|
| [69] |
Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol, 2011, 3(7): a003707.
|
| [70] |
Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell, 2014, 56(1): 55-66. DOI:10.1016/j.molcel.2014.08.019
|
| [71] |
Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep, 2015, 10(1): 103-111. DOI:10.1016/j.celrep.2014.12.002
|
| [72] |
Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell, 2014, 159(1): 134-147. DOI:10.1016/j.cell.2014.09.001
|
| [73] |
Sun X, Wang L, Ding J, et al. Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett, 2016, 590(20): 3510-3516. DOI:10.1002/1873-3468.12440
|
| [74] |
Zhao T, Wang L, Li S, et al. Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors. FEBS Lett, 2017, 591(21): 3660-3669. DOI:10.1002/1873-3468.12868
|
| [75] |
Zeng RF, Zhou JJ, Hu CG, et al. Transcriptome-wide identification and functional prediction of novel and flowering-related circular RNAs from trifoliate orange (Poncirus trifoliata L.Raf.). Planta, 2018, 247(5): 1191-1202. DOI:10.1007/s00425-018-2857-2
|
| [76] |
Wang Y, Gao Y, Zhang H, et al. Genome-wide profiling of circular RNAs in the rapidly growing shoots of moso bamboo (Phyllostachys edulis). Plant Cell Physiol, 2019, 60(6): 1354-1373. DOI:10.1093/pcp/pcz043
|
| [77] |
Tan J, Zhou Z, Niu Y, et al. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci Rep, 2017, 7(1): 8594. DOI:10.1038/s41598-017-08806-0
|
| [78] |
Huang X, Zhang H, Guo R, et al. Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta, 2021, 253(2): 26. DOI:10.1007/s00425-020-03544-6
|
| [79] |
Song Y, Bu C, Chen P, et al. Miniature inverted repeat transposable elements cis-regulate circular RNA expression and promote ethylene biosynthesis, reducing heat tolerance in Populus tomentosa. J Exp Bot, 2021, 72(5): 1978-1994. DOI:10.1093/jxb/eraa570
|
| [80] |
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol, 2014, 32(5): 453-461. DOI:10.1038/nbt.2890
|
| [81] |
Lai XL, Bazin J, Webb S, et al. CircRNAs in plants. Advances in Experimental Medicine and Biology. Singapore: Springer Singapore, 2018: 329-343.
|
| [82] |
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet, 2016, 17(11): 679-692.
|
| [83] |
Guria A, Sharma P, Natesan S, et al. Circular RNAs-the road less traveled. Front Mol Biosci, 2019, 6: 146.
|
| [84] |
Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell, 2016, 165(2): 289-302. DOI:10.1016/j.cell.2016.03.020
|
| [85] |
Tan S, Sun D, Pu W, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer, 2018, 17(1): 138. DOI:10.1186/s12943-018-0887-9
|
| [86] |
Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell, 2019, 176(4): 869-881.e13. DOI:10.1016/j.cell.2018.12.021
|
| [87] |
Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev, 2014, 28(20): 2233-2247. DOI:10.1101/gad.251926.114
|
| [88] |
Kramer MC, Liang DM, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev, 2015, 29(20): 2168-2182. DOI:10.1101/gad.270421.115
|
| [89] |
Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep, 2015, 10(2): 170-177. DOI:10.1016/j.celrep.2014.12.019
|
| [90] |
Zhang Z, Wang H, Wang Y, et al. Whole-genome characterization of chronological age-associated changes in methylome and circular RNAs in moso bamboo (Phyllostachys edulis) from vegetative to floral growth. Plant J, 2021, 106(2): 435-453. DOI:10.1111/tpj.15174
|
| [91] |
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol, 2015, 12(4): 381-388. DOI:10.1080/15476286.2015.1020271
|
| [92] |
Zhang XT, Zhang Y, Wang TY, et al. A comprehensive map of intron branchpoints and lariat RNAs in plants. Plant Cell, 2019, 31(5): 956-973. DOI:10.1105/tpc.18.00711
|
| [93] |
Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants, 2017, 3: 17053. DOI:10.1038/nplants.2017.53
|
| [94] |
Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell, 2015, 160(6): 1125-1134. DOI:10.1016/j.cell.2015.02.014
|
| [95] |
Teplova M, Hafner M, Teplov D, et al. Structure-function studies of STAR family quaking proteins bound to their in vivo RNA target sites. Genes Dev, 2013, 27(8): 928-940. DOI:10.1101/gad.216531.113
|
| [96] |
Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell, 2015, 58(5): 870-885. DOI:10.1016/j.molcel.2015.03.027
|
| [97] |
Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun, 2017, 8: 14741. DOI:10.1038/ncomms14741
|
| [98] |
Aktaş T, Avşar Ilık İ, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature, 2017, 544(7648): 115-119. DOI:10.1038/nature21715
|
| [99] |
Cheng Y, Kato N, Wang W, et al. Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell, 2003, 4(1): 53-66. DOI:10.1016/S1534-5807(02)00399-4
|
| [100] |
Mockler TC, Yu X, Shalitin D, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. PNAS, 2004, 101(34): 12759-12764. DOI:10.1073/pnas.0404552101
|
| [101] |
Rodríguez-Cazorla E, Ripoll JJ, Andújar A, et al. K-homology nuclear ribonucleoproteins regulate floral organ identity and determinacy in Arabidopsis. PLoS Genet, 2015, 11(2): e1004983. DOI:10.1371/journal.pgen.1004983
|
| [102] |
Guan Q, Guan Q, Wen C, et al. A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Mol Plant, 2013, 6(2): 386-395. DOI:10.1093/mp/sss119
|
| [103] |
Jeong IS, Fukudome A, Aksoy E, et al. Regulation of abiotic stress signalling by Arabidopsis C-terminal domain phosphatase-like 1 requires interaction with a K-homology domain-containing protein. PLoS One, 2013, 8(11): e80509. DOI:10.1371/journal.pone.0080509
|
| [104] |
Jiang J, Wang B, Shen Y, et al. The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet, 2013, 9(7): e1003625. DOI:10.1371/journal.pgen.1003625
|
| [105] |
Thatcher LF, Kamphuis LG, Hane JK, et al. The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS One, 2015, 10(5): e0126978. DOI:10.1371/journal.pone.0126978
|
| [106] |
Lorković ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res, 2002, 30(3): 623-635. DOI:10.1093/nar/30.3.623
|
| [107] |
Randles JW, Davies C, Hatta T, et al. Studies on encapsidated viroid-like RNA I.Characterization of velvet tobacco mottle virus. Virology, 1981, 108(1): 111-122. DOI:10.1016/0042-6822(81)90531-6
|
| [108] |
Makino S, Chang MF, Shieh CK, et al. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature, 1987, 329(6137): 343-346. DOI:10.1038/329343a0
|
| [109] |
Rizzetto M. Hepatitis D virus: introduction and epidemiology. Cold Spring Harb Perspect Med, 2015, 5(7): a021576. DOI:10.1101/cshperspect.a021576
|
| [110] |
De La Peña M. Circular RNAs biogenesis in eukaryotes through self-cleaving hammerhead ribozymes. Adv Exp Med Biol, 2018, 1087: 53-63.
|
| [111] |
Olmedo-Velarde A, Navarro B, Hu JS, et al. Novel fig-associated viroid-like RNAs containing hammerhead ribozymes in both polarity strands identified by high-throughput sequencing. Front Microbiol, 2020, 11: 1903. DOI:10.3389/fmicb.2020.01903
|
| [112] |
De La Peña M, Ceprián R, Cervera A. A singular and widespread group of mobile genetic elements: RNA circles with autocatalytic ribozymes. Cells, 2020, 9(12): 2555. DOI:10.3390/cells9122555
|
| [113] |
Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature, 2013, 495(7441): 384-388. DOI:10.1038/nature11993
|
| [114] |
Hong YH, Meng J, Zhang M, et al. Identification of tomato circular RNAs responsive to Phytophthora infestans. Gene, 2020, 746: 144652. DOI:10.1016/j.gene.2020.144652
|
| [115] |
Wang YX, Wang Q, Gao LP, et al. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol Plant, 2017, 161(3): 311-321. DOI:10.1111/ppl.12600
|
| [116] |
Salih H, Wang X, Chen B, et al. Identification, characterization and expression profiling of circular RNAs in the early cotton fiber developmental stages. Genomics, 2021, 113(1 pt 1): 356-365.
|
| [117] |
Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell, 2017, 66(1): 22-37.e9. DOI:10.1016/j.molcel.2017.02.017
|
| [118] |
Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell, 2017, 66(1): 9-21.e7. DOI:10.1016/j.molcel.2017.02.021
|
| [119] |
Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res, 2017, 27(5): 626-641. DOI:10.1038/cr.2017.31
|
| [120] |
Wang X, Chang X, Jing Y, et al. Identification and functional prediction of soybean circRNAs involved in low-temperature responses. J Plant Physiol, 2020, 250: 153188. DOI:10.1016/j.jplph.2020.153188
|
| [121] |
Han Y, Li X, Yan Y, et al. Identification, characterization, and functional prediction of circular RNAs in maize. Mol Genet Genomics, 2020, 295(2): 491-503. DOI:10.1007/s00438-019-01638-9
|
| [122] |
Luo GZ, MacQueen A, Zheng G, et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun, 2014, 5: 5630. DOI:10.1038/ncomms6630
|
| [123] |
Wan Y, Tang K, Zhang D, et al. Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol, 2015, 16: 272. DOI:10.1186/s13059-015-0839-2
|
| [124] |
Pan T, Sun X, Liu Y, et al. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol Biol, 2018, 96(3): 217-229. DOI:10.1007/s11103-017-0684-7
|
| [125] |
Zhu YX, Jia JH, Yang L, et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol, 2019, 19(1): 164. DOI:10.1186/s12870-019-1712-3
|
| [126] |
Zhang P, Fan Y, Sun X, et al. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J, 2019, 98(4): 697-713. DOI:10.1111/tpj.14267
|
| [127] |
Zhao W, Zhang C, Shen X, et al. Characterization of circRNAs associated with resistance to defoliating insectsin soybean. Oil Crop Sci, 2017, 2(001): 23-37.
|
| [128] |
Zhou R, Zhu YX, Zhao J, et al. Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum subspecies brasiliense infection. Int J Mol Sci, 2017, 19(1): E71. DOI:10.3390/ijms19010071
|
| [129] |
Xiang L, Cai C, Cheng J, et al. Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq. Peer J, 2018, 6: e4500. DOI:10.7717/peerj.4500
|
| [130] |
Wang J, Yang Y, Jin L, et al. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol, 2018, 18(1): 104. DOI:10.1186/s12870-018-1332-3
|
| [131] |
Fan J, Quan W, Li GB, et al. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiol, 2020, 182(1): 272-286. DOI:10.1104/pp.19.00716
|
| [132] |
Li Z, Wang S, Cheng J, et al. Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet, 2016, 12(11): e1006422. DOI:10.1371/journal.pgen.1006422
|
| [133] |
Cheng J, Zhang Y, Li Z, et al. A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci China Life Sci, 2018, 61(2): 204-213. DOI:10.1007/s11427-017-9182-3
|
| [134] |
Palukaitis P. Potato spindle Tuber viroid: investigation of the long-distance, intra-plant transport route. Virology, 1987, 158(1): 239-241. DOI:10.1016/0042-6822(87)90260-1
|
| [135] |
Zhu Y, Green L, Woo YM, et al. Cellular basis of potato spindle tuber viroid systemic movement. Virology, 2001, 279(1): 69-77. DOI:10.1006/viro.2000.0724
|
| [136] |
Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J, 2011, 30(21): 4414-4422. DOI:10.1038/emboj.2011.359
|
| [137] |
Park OH, Ha H, Lee Y, et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell, 2019, 74(3): 494-507.e8. DOI:10.1016/j.molcel.2019.02.034
|
| [138] |
Wang X, Lu Z, Gomez A, et al. N6-methyladenosine- dependent regulation of messenger RNA stability. Nature, 2014, 505(7481): 117-120. DOI:10.1038/nature12730
|
| [139] |
Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell, 2019, 177(4): 865-880.e21. DOI:10.1016/j.cell.2019.03.046
|
| [140] |
Jia R, Xiao MS, Li Z, et al. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov, 2019, 5: 45. DOI:10.1038/s41421-019-0113-y
|
| [141] |
Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol, 2007, 17(8): 411-416. DOI:10.1016/j.tcb.2007.06.003
|
| [142] |
Liu J, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol, 2005, 7(12): 1261-1266. DOI:10.1038/ncb1333
|
| [143] |
Guria A, Velayudha Vimala Kumar K, Srikakulam N, et al. Circular RNA profiling by illumina sequencing via template-dependent multiple displacement amplification. Biomed Res Int, 2019, 2019: 2756516.
|
| [144] |
Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet, 2007, 39(8): 1033-1037. DOI:10.1038/ng2079
|
| [145] |
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 2013, 19(2): 141-157. DOI:10.1261/rna.035667.112
|
| [146] |
Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development, 2016, 143(11): 1838-1847. DOI:10.1242/dev.128074
|
| [147] |
Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol, 2017, 14(8): 1035-1045. DOI:10.1080/15476286.2016.1271524
|
| [148] |
Zhou M, Xiao MS, Li Z, et al. New progresses of circular RNA biology: from nuclear export to degradation. RNA Biol, 2021, 18(10): 1365-1373. DOI:10.1080/15476286.2020.1853977
|
| [149] |
Sun HD, Wu ZJ, Liu M, et al. CircRNA may not be "circular". Front Genet, 2021, 12: 633750. DOI:10.3389/fgene.2021.633750
|
| [150] |
Xin R, Gao Y, Gao Y, et al. IsoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat Commun, 2021, 12(1): 266. DOI:10.1038/s41467-020-20459-8
|
| [151] |
Zhang J, Hou L, Zuo Z, et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol, 2021, 39(7): 836-845. DOI:10.1038/s41587-021-00842-6
|
| [152] |
Rahimi K, Venø MT, Dupont DM, et al. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat Commun, 2021, 12(1): 4825. DOI:10.1038/s41467-021-24975-z
|
| [153] |
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol, 2016, 17(4): 205-211. DOI:10.1038/nrm.2015.32
|
| [154] |
Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell, 2018, 71(3): 428-442. DOI:10.1016/j.molcel.2018.06.034
|
| [155] |
Teng F, Cui T, Feng G, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov, 2018, 4: 63.
|
| [156] |
Teng F, Cui T, Gao Q, et al. Artificial sgRNAs engineered for genome editing with new Cas12b orthologs. Cell Discov, 2019, 5: 23. DOI:10.1038/s41421-019-0091-0
|
| [157] |
Li S, Li X, Xue W, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods, 2021, 18(1): 51-59. DOI:10.1038/s41592-020-01011-4
|
| [158] |
Zhou J, Yuan M, Zhao Y, et al. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol J, 2021, 19(6): 1240-1252. DOI:10.1111/pbi.13544
|
| [159] |
Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science, 2017, 358(6366): 1019-1027. DOI:10.1126/science.aaq0180
|
| [160] |
Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature, 2017, 550(7675): 280-284. DOI:10.1038/nature24049
|
 2022, Vol. 38
2022, Vol. 38