中国科学院微生物研究所、中国微生物学会主办
文章信息
- 徐世成, 王鹤冰, 冯俊杰, 向华丰, 吴梦丹, 王志敏, 魏大勇, 张洪成, 汤青林
- XU Shicheng, WANG Hebing, FENG Junjie, XIANG Huafeng, WU Mengdan, WANG Zhimin, WEI Dayong, ZHANG Hongcheng, TANG Qinglin
- 黄瓜霜霉病及寄主抗性机制研究进展
- Cucumber downy mildew and the mechanisms of host resistance: a review
- 生物工程学报, 2022, 38(5): 1724-1737
- Chinese Journal of Biotechnology, 2022, 38(5): 1724-1737
- 10.13345/j.cjb.210513
-
文章历史
- Received: July 6, 2021
- Accepted: September 15, 2021
- Published: September 30, 2021
2. 重庆市农业科学院 蔬菜花卉研究所, 重庆 401329
2. Institute of Vegetables and Flowers, Chongqing Academy of Agricultural Sciences, Chongqing 401329, China
黄瓜(Cucumis sativus L.) 属于葫芦科蔬菜,以幼嫩果实为食[1],种植范围广,是世界十大栽培蔬菜之一[2]。黄瓜霜霉病(downy mildew)由专性卵菌古巴假霜霉菌(Pseudoperonospora cubensis) 引起,属气传性真菌病害,在黄瓜生产中极具破坏性[3-4]。该病原菌在自然条件下不仅侵染黄瓜,还侵染葫芦科大约20个属40个种[5],在黄瓜、甜瓜、南瓜和西瓜上危害尤为严重[6]。1868年在古巴葫芦科作物上首次报道该病原菌。近年来,随着蔬菜种植范围扩大,黄瓜霜霉病发生和传播速度越来越难控制,严重影响全球80多个国家和地区的黄瓜生产[7-8],已于2004年被美国列为黄瓜主要病害[9-10]。
黄瓜霜霉病主要危害黄瓜叶部,也能危害茎和花序,在苗期至成株期间均可发病,尤其黄瓜进入收果期发病较重。当周围环境条件利于黄瓜霜霉病发病时,1–2周内即可导致植株叶片枯死。由于温度、湿度及寄主基因型不同,病害潜伏期一般为4–12 d[11]。病菌对温度、湿度较敏感,高温高湿有利于病害发生和流行。游动孢子囊的数量在高湿条件下迅速增加[12]。同时,侵染的多个病斑结合成大的病斑,导致整片叶坏死、卷缩,整个植株枯萎死亡[13],最终使产量和品质下降甚至绝收。
本文结合作者的科研工作综述了黄瓜霜霉病近年研究进展,并展望了霜霉病的研究方向,为霜霉病的防治、检测和调控、以及抗病新品种选育等提供参考。
1 病原菌防治及检测 1.1 病原菌防治植物病原菌严重威胁着粮油蔬菜安全,它在全球化交流背景下传播更为迅速。霜霉病原菌在黄瓜产区侵染黄瓜会引起严重减产减收,因此急需对该病害及时防治。目前主要的防治方法是综合防治,包括选育抗病品种、生物防治、药剂防治、病原菌预防与早期检测等。其中最好的防治策略是培育优质抗病品种,例如采用杂交法[14]、转基因法[15]、紫外诱变法以及药剂驯化法等创制抗性材料和培育新品种[16]。但是,国内外目前还没有黄瓜霜霉病高抗品种。而且由于不同黄瓜品种的抗病性很可能存在差异,即便同一品种在不同发育时期也有抗性差异,因此培育具有稳定抗性的优质黄瓜新品种显得非常重要但又困难重重。
细菌拮抗剂可作为黄瓜霜霉病的生物防治。针对黄瓜植株田间微环境,综合分析其分离物的遗传多样性、胞外水解酶及次生代谢物特性、拮抗细菌的应答效应等,从而鉴定出5个对黄瓜霜霉病有拮抗性的细菌(短小芽孢杆菌DS22、地衣芽孢杆菌HS10、肠杆菌属DP14、芽孢杆菌属HP4、嗜麦芽窄食单胞菌DS57),其中将DP14、HS10和DS22在田间喷洒黄瓜叶片或者淋根,均可显著抑制黄瓜霜霉病,促进植株生长[17]。
利用离体叶片法筛选发现,多黏类芽孢杆菌P1对黄瓜霜霉病原菌有较强的抑制作用,进一步通过室内测试和田间药效试验表明,多黏类芽孢杆菌P1能减少或替代化学农药的使用,有利于黄瓜霜霉病的绿色防控[18]。最近发现,大豆β-聚赖氨酸和鲶鱼皮粘液p22糖蛋白可以替代杀菌剂,降解黄瓜霜霉病菌游动孢子囊的细胞壁,破坏细胞壁的完整性,从而抑制黄瓜霜霉病[19]。外源褪黑素也可防控蔬菜病害,还可保护苹果等植株免受斑点病侵染、减少损伤数量、减缓发病速率[20]。褪黑素处理对黄瓜霜霉病抗性也有效果,可使黄瓜幼苗的病害指数显著降低。但是,环境因素会较大地影响生物防治效果,而且有益生物培养和繁殖技术难度较高,防治速度也不如药剂防治。
目前病害防治主要以药剂防治为主。黄瓜霜霉病防治效果受到多个因素影响,其中杀菌剂(百菌清和氰霜唑) 效果最佳,能显著抑制病害并使黄瓜产量成倍增加;其次是品种抗性,春季栽培抗病品种Bristol优于感病品种Speedway;但棚架方式对防病效果不显著[21]。然而长期施用单一传统的药剂,会使黄瓜霜霉病产生耐药性。因此有必要研发一些新的杀菌剂和农药,提高黄瓜霜霉病的防控效果。目前开发了以下新的杀菌剂:①嘧啶胺化合物1c杀菌剂,它具有独特抑制复合物NADH氧化还原酶[22];②稀有D-塔格糖新型农药,可以阻断分生孢子的侵染和繁殖,并且具有类似杀真菌剂作用[23];③新衍生物8q杀菌剂,它通过大量腙类化合物与三唑磺酰氯基团偶联合成[24];④氟噻唑吡乙酮杀菌剂[25]。其中嘧啶胺化合物1c和新衍生物8q具有广谱杀菌性,表现出最好的田间杀真菌活性,其药物安全指标的半最大效应浓度EC50 (concentration for 50% of maximal effect) 远高于传统杀菌剂氰霜唑、氟吗啉、霜霉威和吲唑磺菌胺等。这些新型药剂有着显著的广谱杀菌活性和更好的田间药效,具有杀灭黄瓜霜霉病菌的良好潜力。杀菌剂和农药虽然会带来环境污染,但目前仍然应用最为广泛。不可避免地,杀菌剂的频繁使用和误用会导致严重的抗药性。一般而言,病原菌在新杀菌剂使用后两年内可能产生抗药性[26]。
1.2 病原菌检测病原菌的精准检测可以为防病时期、杀菌剂用量等提供指导,有利于高效防治病害和节约杀菌剂。目前国内检测及预防措施包括日光温室黄瓜霜霉病警源追溯系统、冠层温湿度检测预警系统。其中,前者主要利用高光谱成像技术、人工智能技术并结合灾变链式理论、预警理论和植物病害流行学构建而成。国外目前开发了2对不同的特异性引物PcK和PxK,它们基于靶向rDNA基因簇的内部转录间隔区(internal transcribed spacers, ITS) 设计而成,通过双重qPCR (duplex-qPCR) 和HRM (高分辨熔解曲线) 验证,可用于同时检测黄瓜霜霉病和甜瓜白粉病2种病原菌[27]。黄瓜霜霉病菌是专性病原菌,需要活的寄主才能存活和繁殖。由于病原菌DNA样品通常也含有来自其他寄主或附属植物的DNA,因此这种诊断分析方法需具有病原菌高度特异性的引物[28-30]。实现特异性诊断,首先需要阐明霜霉病病原菌的系统发育关系,其次是开发额外的基因组标记。
近期研究表明,便携式DNA测序装置可用于检测空气中传播病原菌的负荷含量和遗传多样性。另外,将高通量DNA测序技术与便携式微流体(DNA测序装置)相结合,以生物传感器的形式来检测多种植物病原菌,可提高对病原菌诊断和生物监测的效率[31-32]。而且,环介导等温扩增(loop-mediated isothermal amplification, LAMP)、交叉引物扩增(crossing priming amplification, CPA) 和SmartAmp (smart amplification process) 与传统PCR方法相比,具有灵敏度高、反应时间短等优点,已被广泛用于多种植物病原菌的检测[33]。这些方法将为黄瓜霜霉病原菌检测提供很好的借鉴。此外,通过已知接种源、远距离大气孢子运输和孢子沉积模块的信息,可准确预测病害暴发的风险,并及时使用杀真菌剂防控病害[34]。与病原菌特异引物扩增鉴定法相比,这类检测方法具有多样化和便捷高效的特点,在侵染初期就可及时准确地监测及预报病情。
综上可知:黄瓜霜霉病的防治策略应该是预防为主,综合防治。首选健壮无病幼苗进行栽植,通过高效简便的病原菌检测方法及时筛选抗病性较强的植株,在黄瓜生长期加强光照,保护地栽培时可短时间迅速提高室内温度抑制病原菌侵染。通过科学施肥、及时在室内通风换气以及减少棚内分生孢子的萌发和入侵,提高抗病性,当病害严重时可使用杀菌剂防治。
2 影响和调控黄瓜霜霉病的因素不同黄瓜品种或不同发育时期对霜霉病的感病程度不同。在侵染霜霉病菌后,感病品种熟性较早而且耐低温,但抗病品种熟性相对较晚且耐热性强。VandenLangenberg和Wehner发现,同一品种不同生育期黄瓜植株对霜霉病的反应不同,与健壮幼龄植株相比,接种霜霉病后老龄成株期发病症状更为严重[35]。另外,不同光照条件也会影响黄瓜霜霉病的发生。在光照与黑暗交替环境下病原菌产生孢子最多,而在持续光照下几乎不会产生游动孢子囊[4]。此外,游动孢子囊在叶面有水滴或水膜时萌发,显症后天数与累积游动孢子囊量之间呈抛物线性相关[36]。黄瓜霜霉病菌具有专性寄生特性,游动孢子囊在活体叶片上产生的数量要多于离体叶片[37]。
此外,黄瓜具有抵抗短期热激的复杂生理机制,从而抑制霜霉病的蔓延发展。热激可使黄瓜霜霉病病情指数降低50%以上,霜霉病菌丝体在45 ℃下暴露数小时后无法存活,说明短期热激可抑制霜霉病蔓延,激活植物抗病性[38-40]。可溶性蛋白和脯氨酸在渗透调节中起重要作用[41],而且两者都与植物抗病性有关[42],在接种病原菌后可溶性蛋白先增后减,但脯氨酸显著下降,外施褪黑素还增加了与这种病原菌相关的酶活性表达,增强抗氧化活性并清除活性氧[43-45]。由此表明,褪黑素可增强霜霉病抗性并且增加黄瓜产量。
植物能识别多种病原菌信号,并快速激活防御机制[46-47]。植物-病原菌相互作用位点快速产生活性氧(reactive oxygen species, ROS),有利于防御反应相关的细胞信号传导[48]、调节过敏反应和程序性细胞死亡[49]。其中酰基高丝氨酸内酯(acyl homoserine lactones, AHLs) 能够抑制病原菌,尤其是长链AHLs能够增强黄瓜植株的防御反应,导致木质素和胼胝质的沉积、防御相关酶激活、活性氧与酚类化合物积累、细胞壁增厚,从而抵御霜霉病菌入侵和危害[50-54]。
3 候选抗病基因的挖掘黄瓜抗病基因的遗传背景较为复杂,多态性标记较难开发,相关育种工作不容易取得突破性进展,而且黄瓜霜霉病抗病优异基因的发掘克隆也会受到极大影响。在美国北卡罗来纳州和新泽西州德尔马瓦半岛,曾经发生严重的黄瓜霜霉病,说明以前鉴定的一些霜霉病(downy mildew) 抗性基因(dm-1、dm-2和dm-3)有可能是水平抗性基因,不能提供足够的病害控制[55]。因此,建立有效的黄瓜种质资源创新体系、发掘更多的垂直抗性优质基因,是促进黄瓜遗传育种的关键。根据抗性蛋白NBS结构域保守基序设计的简并引物,在黄瓜近等基因系IL5211S中分离并且鉴定出28个共4类NBS型基因,命名为黄瓜抗性基因类似物resistance gene analogs (CsRGAs)。实时荧光定量聚合酶链反应(qRT-PCR) 分析表明,黄瓜近等基因系IL5211S中CsRGAs在根、茎和叶中表达水平不同,其中CsRGA23在叶片中的表达显著高于其他3类中的代表性基因CsRGA17、CsRGA22和CsRGA25[56]。在霜霉病菌侵染期间CsRGA23基因被激活,而且茉莉酸、水杨酸、脱落酸和过氧化氢介导的信号转导途径也被显著诱导。由此表明,CsRGA23可能通过这些分子触发的信号通路在黄瓜霜霉病中发挥关键作用。
目前有关植物发育和抗病性之间如何协调的潜在机制仍不清楚。具有碱性螺旋-环-螺旋蛋白(bHLH) 的维管调节因子irregular vasculature patterning (CsIVP) 可结合其他维管发育因子YABBY5 (CsYAB5)、BREVIPEDICELLUS (CsBP) 和AUXIN/INDOLE- ACETIC ACIDS4 (CsAUX4) 的启动子,调控黄瓜维管的系统发育[57]。水杨酸可调节生物病原菌的局部防御反应[58],CsIVP能够与水杨酸信号通路中NIM1-INTERACTING1 (CsNIMIN1) 蛋白直接互作,调控黄瓜霜霉病的抗性。CsIVP转基因干扰植株在接种黄瓜霜霉病后,不管种植在温室还是人工气候箱中都表现出对霜霉病明显的抗性,而且活体营养病原菌的抗性激素水杨酸含量显著增加,水杨酸信号途径中抗性基因PATHOGENESIS-RELATED PROTEIN-1 (PR-1) 的表达也显著上升。由此说明,CsIVP负调节水杨酸形成,并与CsNIMIN1互作来抑制PR-1基因的表达,从而降低黄瓜对病原菌的抗性。
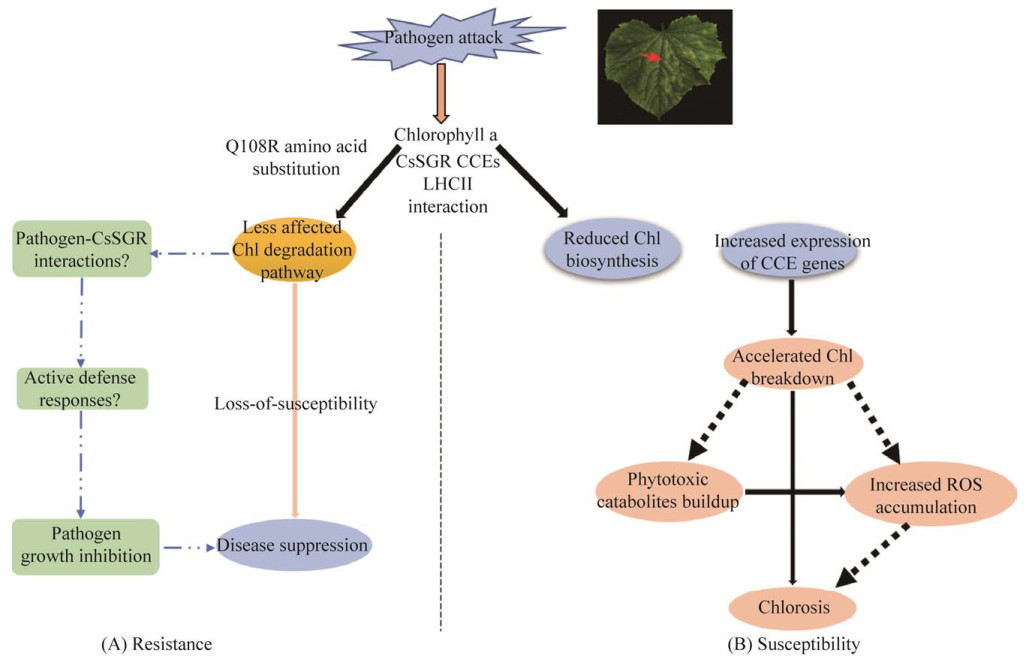
此外黄瓜的滞绿基因Stay Green Gene (CsSGR) 是一个易感性(loss-of-susceptibility)丧失的寄主抗性基因,也是霜霉病抗性位点dm1/psl/cla基因座的候选基因。黄瓜品种Gy14的抗病性是由于CsSGR蛋白中的Q108R氨基酸突变所致[59]。不同抗感品种中叶绿素降解途径相关基因的表达存在差异,黄瓜CsSGR和拟南芥AtSGA1在叶绿素降解中起着重要的调节作用,这也可能与抗病性有关[60-61]。植物通过激活不同的防御反应来对抗多种病原菌[62-65],活性氧常常作为信号分子在抵抗病原菌侵染的防御反应中发挥作用[66-70]。突变的CsSGR蛋白与叶绿素分解代谢酶chlorophyll catabolism enzymes (CCEs) 和光系统Ⅱ捕光色素蛋白复合体light- harvesting complex subunits of photosystem Ⅱ (LHCII) 相互作用,调节抗性株系中活性氧和叶绿素(Chl),从而减轻损伤。在易感株系中,由于病原菌侵染使CCEs基因表达增加、叶绿素生物合成减少、活性氧增加、植物毒性分解代谢物堆积,从而导致叶片黄化(图 1)。
植物在与病原菌长期斗争中逐渐进化并形成复杂的防御机制[71]。在病原菌侵染后,植物会改变蛋白质水平、产生信号级联反应和生物防御效应。基因组学研究可以全面系统地解析病原菌的应答效应、细胞代谢和其他过程[72]。蛋白质组学研究也逐渐用于黄瓜抗性新基因的挖掘以及黄瓜选择性育种[73]。
利用双向电泳(two-dimensional electrophoresis, 2-DE) 以及基质辅助激光解吸电离串联飞行时间质谱(MALDI-TOF MS),已鉴定出黄瓜霜霉病响应蛋白90个[74]。这些差异积累蛋白(DAPs) 广泛参与代谢、运输、蛋白折叠、蛋白水解、细胞增殖、光合作用、葡萄糖代谢、翻译延伸和损伤等过程。当霜霉病菌侵染黄瓜叶片后,超氧化物歧化酶(superoxide dismutase, SOD) 和过氧化物酶(peroxidase, POD) 活性较对照明显升高[75]。通过KEGG分析发现,DAPs主要聚集于固碳、氧化磷酸化、氨基酸生物合成等多种途径[74-75]。另外,光合作用在植物防御中起着重要作用。例如,甘油醛-3-磷酸脱氢酶(GAPDH) 在固碳过程中受到病菌侵染的影响[76]。同样热休克蛋白作为差异积累蛋白在黄瓜霜霉病抗性和胁迫中发挥作用[71]。此外,在黄瓜霜霉病侵染黄瓜抗病品种“ZJ”和感病品种“SDG”的过程中,许多代谢途径(例如“ZJ”中硒化合物代谢和“SDG”中淀粉和蔗糖代谢) 发生了显著变化。许多与发病机制相关的蛋白质如几丁质酶、过氧化物酶、抗性蛋白也发生了变化[77]。这表明黄瓜霜霉病的抗性受到复杂的网络调控。
通过转录组分析已鉴定出多个黄瓜霜霉病响应相关基因。其中,利用抑制性消减杂交(SSH) 构建了正向消减文库(FSL) 和反向消减文库(RSL),并分别分离出1 416个和1 128个重组克隆。对优先表达的重组克隆进行差异筛选,分别鉴定出58个来自FSL和29个来自RSL的独特表达序列标签。它们分别属于信号转导、细胞防御、细胞周期、蛋白质结合和代谢等植物防御途径[78]。这有利于黄瓜霜霉病抗性机制的深入理解和进一步研究。
利用基因组分析发现,黄瓜WRKY家族中至少有12个成员(CsWRKY 10/14/19/27/28/32/ 35/46/50/52/59/61) 对霜霉病和白粉病的侵染有反应[79-80]。WRKY转录因子可以结合到目标基因的启动子,调节基因的表达,影响植物生长发育、应答生物或非生物胁迫。此外,WRKY转录因子还参与激素等信号转导。其中CsWRKY50转基因株系能增强霜霉病抗性,并且可能通过水杨酸和茉莉酸途径参与调控黄瓜霜霉病[81]。锌指同源结构域(ZF-HD) 蛋白编码一个植物特异性转录因子家族,调节植物生长发育、激活或抑制靶基因对非生物/生物胁迫的反应。Lai等[82]首次在黄瓜基因组中鉴定出13个ZF-高密度基因(命名为CsMIF1-CsMIF3和CsZHD1-CsZHD10),它们不均匀地分布在6条染色体上,其中CsZHD1–3、CsZHD6、CsZHD8和CsZHD10在接种霜霉病菌后表达显著下调,而CsZHD7的表达显著增加。此外,CsZHD4在接种霜霉病后差异表达。
VQ蛋白广泛存在于高等植物,也能够调控植物生长发育、应答病原菌胁迫[83-84],部分VQ蛋白还可以与WRKY转录因子互作。例如,拟南芥AtVQ21/MKS1能够结合到MAPK4蛋白,参与水杨酸信号途径,增强植株对丁香假单胞菌的抗性。此外AtVQ21/MKS1也能负向调节茉莉酸信号途径,减弱植株对灰霉病菌的抗性[85-87]。AtVQ23 (SIB1) 和AtVQ16 (SIB2) 的转录表达受到灰霉病菌的强烈诱导,且通过与WRKY33相互作用,增强植株对灰霉病的抗性[88]。这些研究结果对黄瓜霜霉病抗性机制研究具有很好的借鉴作用。
前述也表明:起初发现至少有1个单隐性基因dm控制着霜霉病,后来发现3个其他dm基因或甚至更多的隐性基因也参与其中。另外,黄瓜霜霉病菌侵染可显著诱导CsRGA23、黄瓜热激蛋白(HSP45.9) 基因和CsWRKY50基因表达,参与病原菌的防御反应,可以作为抗病候选基因。但是CsZHD1、CsZHD2、CsZHD3、CsZHD5和CsZHD9的抗性机制还有待进一步鉴定。在黄瓜霜霉病抗性机制研究中,越来越多的防御相关基因和蛋白被发掘。因此,有必要从组学角度研究更多的黄瓜遗传资源,挖掘更多潜在的霜霉病抗病基因和蛋白质,以促进黄瓜遗传改良和抗性育种。
5 黄瓜霜霉病QTL连锁标记开发数量性状位点(quantitative trait locus, QTL)定位是理解表型变异基因组的基本方法,可为作物育种中候选基因的图位克隆和标记辅助选择奠定基础。Wang等[89]在黄瓜染色体2、4、5和6上分别鉴定了4个稳定的QTL抗霜霉病基因,通过QTL作图分析了霜霉病抗性的遗传基础。Yoshioka等对重组近交系(recombinant inbred line, RIL) 群体进行QTL分析,发现10个QTL被绘制在1、3、5、6和7号染色体上[90]。此外,Vandenlangenberg在4号和5号染色体上检测到霜霉病抗性的3个候选(数量性状位点) QTL[91]。由此表明:黄瓜霜霉病抗性受复杂的遗传系统控制。Li等在4号染色体上检测到1个主效基因:dm4.1,认为黄瓜对于霜霉病的抗性不仅取决于寄主的遗传抗性,还取决于影响QTL检测的其他因素(例如,检测方法、环境条件、病原菌种类和植物生长阶段)[92]。最近,Szczechura等从黄瓜PI 197085的第5号染色上鉴定出3个霜霉病抗性基因(DM1、DM2、DM3)[93]。由此表明:大规模分离分析(BSA)和后代测序(NGS) 技术相结合是一种快速、低成本且高效的QTL定位方法。
Win等将NGS辅助下的BSA应用于黄瓜的QTL抗性作图,检测到5个QTL (dm2.2、dm4.1、dm5.1、dm5.2和dm6.1),其中dm2.2对霜霉病抗性的影响最大[94]。利用F2的常规QTL分析确定dm 2.2和dm 5.2为主要QTL,dm4.1为次要QTL,但不能检测dm5.1和dm6.1。基于F3群体的QTL方法在黄瓜2号染色体上鉴定了新的位点dm2.1。
Wang等利用SSR标记发现,有422个具有多态性(27.5%),其中将312个均匀分布在7条染色体上的标记用于检测抗病和感病群体,发现有19个标记在群体之间具有多态性[95]。这19个标记分布在5条染色体上(染色体1、3、4、5和6)。单标记分析显示,19个标记中有16个与霜霉病抗性显著相关。所检测到的dm5.1、dm5.2和dm5.3是新发现的霜霉病抗性标记。霜霉病抗性(DMR) 和白粉病抗性(PMR) 可能存在重叠的标记,但是它们之间的遗传关系仍不清楚[96]。在感病品种“长春密刺”和IL52的杂交品种中,发现1个定位到5号染色体上大约468 kb的单隐性基因pm。结合连锁分析和极端性状混池重测序(BSA-seq),pm与主效应DMR QTL dm5.2共定位。该共定位位点pm/dm5.2对白粉病具有完全抗性,对霜霉病具有部分抗性;在dm5.2中鉴定出7个霜霉病候选基因。
6 黄瓜霜霉病抗病基因利用及新品种选育最近对不同来源或生态型的黄瓜核心种质(CG) 重测序,并进行全基因组关联分析(GWAS),在黄瓜7条染色体上共检测到18个均匀分布的位点[97]。这18个位点中只有6个位点(dmG1.4、dmG4.1、dmG4.3、dmG5.2、dmG7.1和dmG7.2) 对霜霉病具有稳定的抗性作用。另外,与QTL共定位位点16个,dmg2.1和dmg7.1是新发现的2个位点。这为更多候选基因的挖掘及抗霜霉病育种提供了借鉴。例如,编码NBS的抗性基因CsRGA23、热激蛋白HSP45.9基因、CsWRKY50等作为抗病候选基因均参与了霜霉病的防御反应。目前尚缺乏这些转基因的黄瓜新品种,但它们具有黄瓜抗霜霉病的分子育种潜力,这些抗病候选基因将来有望应用于黄瓜分子辅助育种。
在抗病品种选育方面,利用中等抗性的黄瓜品种马克特莫尔97和象牙皇后以及康奈尔大学开发的品种,培育出了具有优异抗病性的黄瓜品系。康奈尔育种品系DMR-NY 264具有最高水平的黄瓜霜霉病抗性,DMR-NY 264品系的植株在没有施用杀真菌剂的情况下结出优质的黄瓜果实[98],这些优质的品种选育也可以为我国黄瓜育种提供参考和借鉴。
此外,已通过杂交育种成功选育出一批抗霜霉病的优质黄瓜新品种:“中农1号”“津杂1号”“津杂4号”“津春4号”等。它们在品质、产量和抗病性方面均显著优于对照品种。华南型黄瓜有着良好的选育前景,但目前华北密刺型黄瓜仍备受育种者关注。就品种数量而言,华南型黄瓜在2020年首次超过华北型。它们均为常规育种技术选育而成,目前利用转基因等分子育种技术直接选育的黄瓜抗霜霉病品种还未见报道,这可能与霜霉病抗性的分子调控机制的复杂性、转基因安全性等因素有关。
目前通过引种、选种或杂交育种等手段,选育出了高产抗病的黄瓜新品种。利用抗病品种来防治病害是最经济、有效和安全的措施,在现代农业科学中,随着对分子生物学和遗传学深入研究以及农业技术的不断开发,抗病分子育种具有广阔的发展前景。但并非所有病害都能在短期内培育出抗病品种,许多病害还需采用其他防治技术进行控制。由于科学技术水平或实验手段的限制,对专性寄生物(霜霉菌、植物病毒和类病毒、类菌原体和类细菌等),目前无法在合成培养基上培养成功来获得纯培养,这也限制了许多生物学性状的深入研究。
7 展望黄瓜霜霉病严重影响世界黄瓜产业的发展,不少研究者在黄瓜霜霉病的病原菌检测及防治、影响因素、抗病候选基因发掘以及基于蛋白质组和基因组的分析等方面取得了一些进展。随着生物技术的不断发展,寄主诱导的基因沉默、基因编辑等可为抗病基因功能及机制研究提供技术支撑。有关黄瓜霜霉病方面的研究还不够深入,寄主与病原菌之间关系复杂的作用机制尚待进一步阐释。
由于缺少统一的鉴定方法以及黄瓜霜霉病菌生理小种的不断分化,目前小种的划分存在分歧[99-102]。黄瓜霜霉病菌表现出寄主专化性,可以根据它们与特定瓜类寄主的类型将具有不同毒性的病原菌进行致病力分类。然而,在霜霉病菌中寄主特化的遗传基础尚不清楚,亟待深入研究加以解决。
首先应该加强黄瓜霜霉病菌致病机理的研究和认识,其次。由于霜霉病抗性基因功能验证并不多,对于抗病基因的挖掘还需大量工作。在病原菌与抗病植株的相互作用过程中,是否有特定物质通过识别来激活和参与植物自身防御反应,在抗病过程中是否涉及额外的调节因子或信号通路等问题都是未知的。
长链非编码RNAs (lncRNAs) 和miRNAs在植物发育和免疫中发挥重要作用。目前,Nie等[103]分析了2个黄瓜抗性品种(T12-19和12-85) 和易感品种(91-112) 在接种白粉病病菌后相关的mRNAs和长链非编码RNAs,发现参与防御反应的差异表达的基因在抗性品种中比在易感品种中多。然而在黄瓜霜霉病中关于长链非编码mRNA参与植株抗病途径方面还缺乏相关报道,目前有且仅有关于白粉病的报道。
此外,WRKY家族相关基因的功能及其黄瓜霜霉病调控机制也值得深入研究[79]。目前发现,WRKY转录因子可能与病程相关基因非表达子1 (nonexpressor of pathogenesis- related genes 1, NPR1) 互作,通过水杨酸信号途径调控黄瓜霜霉病抗性[104]。
黄瓜新品种的抗病性对于防控霜霉病具有重要意义,为满足生产和消费需要,目前我们已培育出“燕白” “燕青” “新燕095”等一系列适于本地的黄瓜品种。为了深入挖掘优质种质,可以结合QTL-seq、转录组测序等鉴定出抗黄瓜霜霉病的InDel、SNP或SSR等分子标记以及相关基因。我们近期利用BSA法构建极端基因池,获得了黄瓜霜霉病的SSR和SRAP分子标记,将应用于黄瓜霜霉病抗性亲本筛选、回交群体的早期分子辅助选择、杂交组合及自交后代的苗期抗霜霉病鉴定。此外,利用基因敲除和转基因技术,定向编辑黄瓜滞绿基因Stay Green Gene (CsSGR)、WRKY家族因子、以及水杨酸信号途径核心元件等,创制抗霜霉病黄瓜育种材料,培育黄瓜抗病优质新品种,这也值得深入研究。
| [1] |
刘东. 黄瓜霜霉病及棒孢叶斑病双抗性分子机制的研究[D]. 哈尔滨: 东北农业大学, 2017. Liu D. Study on molecular mechanism of double diseases resistance to cucumber downy mildew and target spot[D]. Harbin: Northeast Agricultural University, 2017 (in Chinese). |
| [2] |
熊艳, 王鹤冰, 向华丰, 等. 黄瓜霜霉病研究进展. 中国农学通报, 2016, 32(1): 130-135. Xiong Y, Wang HB, Xiang HF, et al. Research progress of cucumber powery mildew. Chin Agric Sci Bull, 2016, 32(1): 130-135 (in Chinese). |
| [3] |
Göker M, Voglmayr H, Riethmüller A, et al. How do obligate parasites evolve.A multi-gene phylogenetic analysis of downy mildews. Fungal Genet Biol, 2007, 44(2): 105-122. DOI:10.1016/j.fgb.2006.07.005
|
| [4] |
Savory EA, Granke LL, Quesada-Ocampo LM, et al. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol Plant Pathol, 2011, 12(3): 217-226. DOI:10.1111/j.1364-3703.2010.00670.x
|
| [5] |
石延霞, 李宝聚, 刘学敏. 黄瓜霜霉病研究进展. 东北农业大学学报, 2002, 33(4): 391-395. Shi YX, Li BJ, Liu XM. The study of cucumber downy mildew. J Northeast Agric Univ, 2002, 33(4): 391-395 (in Chinese). DOI:10.3969/j.issn.1005-9369.2002.04.015 |
| [6] |
Lebeda A, Widrlechner MP. A set of Cucurbitaceae taxa for differentiation of Pseudoperonospora cubensis pathotypes. Z Pflanzenk Pflanzen, 2003, 110(4): 337-349.
|
| [7] |
Thakur RP, Mathur K. Downy mildews of India. Crop Prot, 2002, 21(4): 333-345. DOI:10.1016/S0261-2194(01)00097-7
|
| [8] |
朱金英, 王友平, 郭平银, 等. 黄瓜霜霉病研究进展. 北方园艺, 2008(4): 74-78. Zhu JY, Wang YP, Guo PY, et al. Progress of study on downy mildew in cucumber. North Hortic, 2008(4): 74-78 (in Chinese). |
| [9] |
Holmes G, Thomas C. 2009. The history and re-emergence of cucurbit downy mildew. Phytopathology, 99(6): S171-S171.
|
| [10] |
Holmes GJ, Main CE, Keever ZT. Cucurbit downy mildew: a unique pathosystem for disease forecasting//Spencer-Phillips P, Jeger P. ADVANCES in DOWNY MILDEW RESEARCH. Volume 2. Dordrecht: Springer, 2004: 69-80.
|
| [11] |
Lebeda A, Cohen Y. Cucurbit downy mildew (Pseudoperonospora cubensis)—biology, ecology, epidemiology, host-pathogen interaction and control. Eur J Plant Pathol, 2011, 129(2): 157-192. DOI:10.1007/s10658-010-9658-1
|
| [12] |
石延霞, 李宝聚, 刘学敏. 黄瓜霜霉病菌致病作用与两种细胞壁降解酶关系初探. 园艺学报, 2003, 30(4): 465-466. Shi YX, Li BJ, Liu XM. The relation of pathogenesis action by Pseudoperonospora cubensis and two sort of zymin in cucumber. Acta Hortic Sin, 2003, 30(4): 465-466 (in Chinese). DOI:10.3321/j.issn:0513-353X.2003.04.023 |
| [13] |
Oerke EC, Steiner U, Dehne HW, et al. Thermal imaging of cucumber leaves affected by downy mildew and environmental conditions. J Exp Bot, 2006, 57(9): 2121-2132. DOI:10.1093/jxb/erj170
|
| [14] |
曹清河. 黄瓜抗霜霉病异源易位系选育、相关基础研究及育种应用[D]. 南京: 南京农业大学, 2006. Cao QH. Reasearch on cucumber alien translocation line possessing resistance to downy mildew and its application in cucumber breeding[D]. Nanjing: Nanjing Agricultural University, 2006 (in Chinese). |
| [15] |
Yin Z, Hennig J, Szwacka M, et al. Tobacco PR-2d promoter is induced in transgenic cucumber in response to biotic and abiotic stimuli. J Plant Physiol, 2004, 161(5): 621-629. DOI:10.1078/0176-1617-00737
|
| [16] |
王岩, 冯明鸣, 刘鹏飞, 等. 黄瓜霜霉病菌对烯肟菌酯敏感性及其抗药性突变体生物学性状研究. 植物病理学报, 2005, 35(S1): 111-112. Wang Y, Feng MM, Liu PF, et al. Detection on sensitivity of Pseudoperonospora cubensis to enostrobilurin and characterization of its laboratory resistant mutants. Acta Phytopathol Sin, 2005, 35(S1): 111-112 (in Chinese). |
| [17] |
Zheng L, Gu C, Cao J, et al. Selecting bacterial antagonists for cucurbit downy mildew and developing an effective application method. Plant Dis, 2018, 102(3): 628-639. DOI:10.1094/PDIS-01-17-0058-RE
|
| [18] |
叶乃玮, 王承芳, 干华磊, 等. 多黏类芽胞杆菌Paenibacillus polymyxa菌株P1防治黄瓜霜霉病的研究. 植物保护, 2021, 47(2): 271-275. Ye NW, Wang CF, Gan HL, et al. Control effect of Paenibacillus polymyxa strain P1 against cucumber downy mildew. Plant Prot, 2021, 47(2): 271-275 (in Chinese). |
| [19] |
Atallah OO, Osman A, Ali MA, et al. Soybean β-conglycinin and catfish cutaneous mucous p22 glycoproteins deteriorate sporangial cell walls of Pseudoperonospora cubensis and suppress cucumber downy mildew. Pest Manag Sci, 2021, 77(7): 3313-3324. DOI:10.1002/ps.6375
|
| [20] |
Yin L, Wang P, Li M, et al. Exogenous melatonin improves Malus resistance to Marssonina apple blotch. J Pineal Res, 2013, 54(4): 426-434. DOI:10.1111/jpi.12038
|
| [21] |
Keinath AP. Integrated management of downy mildew on slicing cucumber with fungicides and host resistance but not trellising. Plant Dis, 2019, 103(10): 2592-2598. DOI:10.1094/PDIS-02-19-0323-RE
|
| [22] |
Guan AY, Wang MG, Yang JL, et al. Discovery of a new fungicide candidate through lead optimization of pyrimidinamine derivatives and its activity against cucumber downy mildew. J Agric Food Chem, 2017, 65(49): 10829-10835. DOI:10.1021/acs.jafc.7b03898
|
| [23] |
Mochizuki S, Fukumoto T, Ohara T, et al. The rare sugar D-tagatose protects plants from downy mildews and is a safe fungicidal agrochemical. Commun Biol, 2020, 3(1): 423. DOI:10.1038/s42003-020-01133-7
|
| [24] |
Li YT, Lin J, Yao WQ, et al. Discovery of a new fungicide by screening triazole sulfonylhydrazone derivatives and its downy mildew inhibition in cucumber. J Heterocyclic Chem, 2020, 57(5): 2128-2138. DOI:10.1002/jhet.3932
|
| [25] |
Salas SE, Shepherd CP, Ngugi HK, et al. Disease control attributes of oxathiapiprolin fungicides for management of cucurbit downy mildew. Plant Dis, 2019, 103(11): 2812-2820. DOI:10.1094/PDIS-02-19-0396-RE
|
| [26] |
Jian W, He D, Xi P, et al. Synthesis and biological evaluation of novel fluorine-containing stilbene derivatives as fungicidal agents against phytopathogenic fungi. J Agric Food Chem, 2015, 63(45): 9963-9. DOI:10.1021/acs.jafc.5b04367
|
| [27] |
Bandamaravuri KB, Nayak AK, Bandamaravuri AS, et al. Simultaneous detection of downy mildew and powdery mildew pathogens on Cucumis sativus and other cucurbits using duplex-qPCR and HRM analysis. AMB Express, 2020, 10(1): 135. DOI:10.1186/s13568-020-01071-x
|
| [28] |
Thines M, Telle S, Ploch S, et al. Identity of the downy mildew pathogens of basil, Coleus, and sage with implications for quarantine measures. Mycol Res, 2009, 113(5): 532-540. DOI:10.1016/j.mycres.2008.12.005
|
| [29] |
Derevnina L, Chin-Wo-Reyes S, Martin F, et al. Genome sequence and architecture of the tobacco downy mildew pathogen Peronospora tabacina. Mol Plant Microbe Interact, 2015, 28(11): 1198-1215. DOI:10.1094/MPMI-05-15-0112-R
|
| [30] |
Sharma R, Xia X, Cano LM, et al. Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics, 2015, 16: 741. DOI:10.1186/s12864-015-1904-7
|
| [31] |
Nezhad AS. Future of portable devices for plant pathogen diagnosis. Lab Chip, 2014, 14(16): 2887-2904. DOI:10.1039/C4LC00487F
|
| [32] |
Ray M, Ray A, Dash S, et al. Fungal disease detection in plants: traditional assays, novel diagnostic techniques and biosensors. Biosens Bioelectron, 2017, 87: 708-723. DOI:10.1016/j.bios.2016.09.032
|
| [33] |
Mahaffee WF. Use of airborne inoculum detection for disease management decisions. Detection and Diagnostics of Plant Pathogens. Dordrecht: Springer Netherlands, 2014: 39-54.
|
| [34] |
Neufeld KN, Keinath AP, Gugino BK, et al. Predicting the risk of cucurbit downy mildew in the eastern United States using an integrated aerobiological model. Int J Biometeorol, 2018, 62(4): 655-668. DOI:10.1007/s00484-017-1474-2
|
| [35] |
VandenLangenberg KM, Wehner TC. Downy mildew disease progress in resistant and susceptible cucumbers tested in the field at different growth stages. HortScience, 2016, 51(8): 984-988. DOI:10.21273/HORTSCI.51.8.984
|
| [36] |
Cohen Y, Rotem J. Field and growth chamber approach to epidemiology of Pseudoperonospora cubensis on cucumbers. Phytopathology, 1971, 61(6): 736. DOI:10.1094/Phyto-61-736
|
| [37] |
Salehi F, Lacroix R, Wade KM. Effects of learning parameters and data presentation on the performance of backpropagation networks for milk yield prediction. Trans ASAE, 1998, 41(1): 253-259. DOI:10.13031/2013.17144
|
| [38] |
Cohen Y, Rubin AE. Daytime solar heating controls downy mildew Peronospora belbahrii in sweet basil. PLoS One, 2015, 10(5): e0126103. DOI:10.1371/journal.pone.0126103
|
| [39] |
Sato T, Kubo M. Reducing the need for chemical spraying of summer greenhouse cucumber: heat-shock controls disease and insect damage. Acta Hortic, 2002(588): 165-170.
|
| [40] |
Ding X, Jiang Y, Hao T, et al. Effects of heat shock on photosynthetic properties, antioxidant enzyme activity, and downy mildew of cucumber (Cucumis sativus L.). PLoS One, 2016, 11(4): e0152429. DOI:10.1371/journal.pone.0152429
|
| [41] |
Zhao XX, Huang LK, Zhang XQ, et al. Effects of heat acclimation on photosynthesis, antioxidant enzyme activities, and gene expression in orchardgrass under heat stress. Molecules, 2014, 19(9): 13564-13576. DOI:10.3390/molecules190913564
|
| [42] |
Meng JF, Xu TF, Wang ZZ, et al. The ameliorative effects of exogenous melatonin on grape cuttings under water-deficient stress: antioxidant metabolites, leaf anatomy, and chloroplast morphology. J Pineal Res, 2014, 57(2): 200-212. DOI:10.1111/jpi.12159
|
| [43] |
Galano A, Tan DX, Reiter RJ. Melatonin as a natural ally against oxidative stress: a physicochemical examination. J Pineal Res, 2011, 51(1): 1-16. DOI:10.1111/j.1600-079X.2011.00916.x
|
| [44] |
Zhang N, Zhao B, Zhang HJ, et al. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J Pineal Res, 2013, 54(1): 15-23. DOI:10.1111/j.1600-079X.2012.01015.x
|
| [45] |
Sun YK, Liu ZY, Lan GP, et al. Effect of exogenous melatonin on resistance of cucumber to downy mildew. Sci Hortic, 2019, 255: 231-241. DOI:10.1016/j.scienta.2019.04.057
|
| [46] |
Beckers GJ, Conrath U. Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol, 2007, 10(4): 425-431. DOI:10.1016/j.pbi.2007.06.002
|
| [47] |
Conrath U, Beckers GJM, Langenbach CJG, et al. Priming for enhanced defense. Annu Rev Phytopathol, 2015, 53(1): 97-119. DOI:10.1146/annurev-phyto-080614-120132
|
| [48] |
Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol, 1997, 48(1): 251-275. DOI:10.1146/annurev.arplant.48.1.251
|
| [49] |
De Pinto MC, Locato V, De Gara L. Redox regulation in plant programmed cell death. Plant Cell Environ, 2012, 35(2): 234-244. DOI:10.1111/j.1365-3040.2011.02387.x
|
| [50] |
Pazarlar S, Cetinkaya N, Bor M, et al. N-acyl homoserine lactone-mediated modulation of plant growth and defense against Pseudoperonospora cubensis in cucumber. J Exp Bot, 2020, 71(20): 6638-6654. DOI:10.1093/jxb/eraa384
|
| [51] |
Schikora A, Schenk ST, Stein E, et al. N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol, 2011, 157(3): 1407-1418. DOI:10.1104/pp.111.180604
|
| [52] |
Schenk ST, Stein E, Kogel KH, et al. Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal Behav, 2012, 7(2): 178-181. DOI:10.4161/psb.18789
|
| [53] |
West J, Kimber R. Innovations in air sampling to detect plant pathogens. Ann Appl Biol, 2015, 166(1): 4-17. DOI:10.1111/aab.12191
|
| [54] |
Schenk ST, Schikora A. AHL-priming functions via oxylipin and salicylic acid. Front Plant Sci, 2014, 5: 784.
|
| [55] |
Horejsi T, Staub JE, Thomas C. Linkage of random amplified polymorphic DNA markers to downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica, 2000, 115(2): 105-113. DOI:10.1023/A:1003942228323
|
| [56] |
Wan H, Zhao Z, Malik AA, et al. Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC Plant Biol, 2010, 10: 186. DOI:10.1186/1471-2229-10-186
|
| [57] |
Yan SS, Ning K, Wang ZY, et al. CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol, 2020, 18(3): e3000671. DOI:10.1371/journal.pbio.3000671
|
| [58] |
Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol, 2009, 47: 177-206. DOI:10.1146/annurev.phyto.050908.135202
|
| [59] |
Wang Y, Tan J, Wu Z, et al. STAYGREEN, STAY HEALTHY: a loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol, 2019, 221(1): 415-430. DOI:10.1111/nph.15353
|
| [60] |
Shimoda Y, Ito H, Tanaka A. Arabidopsis STAY-GREEN, mendel's green cotyledon gene, encodes magnesium-dechelatase. Plant Cell, 2016, 28(9): 2147-2160. DOI:10.1105/tpc.16.00428
|
| [61] |
Kuai BK, Chen JY, Hörtensteiner S. The biochemistry and molecular biology of chlorophyll breakdown. J Exp Bot, 2018, 69(4): 751-767. DOI:10.1093/jxb/erx322
|
| [62] |
Thomma BP, Eggermont K, Penninckx IA, et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. PNAS, 1998, 95(25): 15107-15111. DOI:10.1073/pnas.95.25.15107
|
| [63] |
Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol, 2005, 43: 205-227. DOI:10.1146/annurev.phyto.43.040204.135923
|
| [64] |
Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. PNAS, 2007, 104(47): 18842-18847. DOI:10.1073/pnas.0708139104
|
| [65] |
Mengiste T. Plant immunity to necrotrophs. Annu Rev Phytopathol, 2012, 50: 267-294. DOI:10.1146/annurev-phyto-081211-172955
|
| [66] |
Mach JM, Castillo AR, Hoogstraten R, et al. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. PNAS, 2001, 98(2): 771-776. DOI:10.1073/pnas.98.2.771
|
| [67] |
Hirashima M, Tanaka R, Tanaka A. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol, 2009, 50(4): 719-729. DOI:10.1093/pcp/pcp035
|
| [68] |
Mur LA, Aubry S, Mondhe M, et al. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol, 2010, 188(1): 161-174. DOI:10.1111/j.1469-8137.2010.03377.x
|
| [69] |
Ishiga Y, Uppalapati SR, Gill US, et al. Transcriptomic and metabolomic analyses identify a role for chlorophyll catabolism and phytoalexin during Medicago nonhost resistance against Asian soybean rust. Sci Rep, 2015, 5: 13061. DOI:10.1038/srep13061
|
| [70] |
Serrano I, Audran C, Rivas S. Chloroplasts at work during plant innate immunity. J Exp Bot, 2016, 67(13): 3845-3854. DOI:10.1093/jxb/erw088
|
| [71] |
Rakwal R, Komatsu S. Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis, 2000, 21(12): 2492-2500. DOI:10.1002/1522-2683(20000701)21:12<2492::AID-ELPS2492>3.0.CO;2-2
|
| [72] |
Pang Z, Srivastava V, Liu X, et al. Quantitative proteomics links metabolic pathways to specific developmental stages of the plant-pathogenic oomycete Phytophthora capsici. Mol Plant Pathol, 2017, 18(3): 378-390. DOI:10.1111/mpp.12406
|
| [73] |
Gong B, Nie W, Yan Y, et al. Unravelling cadmium toxicity and nitric oxide induced tolerance in Cucumis sativus: insight into regulatory mechanisms using proteomics. J Hazard Mater, 2017, 336: 202-213. DOI:10.1016/j.jhazmat.2017.04.058
|
| [74] |
Nostar O, Ozdemir F, Bor M, et al. Combined effects of salt stress and cucurbit downy mildew (Pseudoperospora cubensis Berk. and Curt. Rostov.) infection on growth, physiological traits and antioxidant activity in cucumber (Cucumis sativus L.) seedlings. Physiol Mol Plant Pathol, 2013, 83: 84-92. DOI:10.1016/j.pmpp.2013.05.004
|
| [75] |
Kim MD, Kim YH, Kwon SY, et al. Overexpression of 2-cysteine peroxiredoxin enhances tolerance to methyl viologen-mediated oxidative stress and high temperature in potato plants. Plant Physiol Biochem, 2011, 49(8): 891-897. DOI:10.1016/j.plaphy.2011.04.001
|
| [76] |
Zhang P, Zhu YQ, Shen CJ, et al. Proteome analysis of cucumber responses to Pseudoperonospora cubensis infection. J Plant Pathol, 2019, 101(4): 917-925. DOI:10.1007/s42161-019-00290-x
|
| [77] |
Zhang P, Zhu YQ, Luo XJ, et al. Comparative proteomic analysis provides insights into the complex responses to Pseudoperonospora cubensis infection of cucumber (Cucumis sativus L.). Sci Rep, 2019, 9: 9433. DOI:10.1038/s41598-019-45111-4
|
| [78] |
Li JW, Liu J, Zhang H, et al. Identification and transcriptional profiling of differentially expressed genes associated with resistance to Pseudoperonospora cubensis in cucumber. Plant Cell Rep, 2011, 30(3): 345-357. DOI:10.1007/s00299-010-0959-9
|
| [79] |
Chen C, Chen X, Han J, et al. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol, 2020, 20(1): 443. DOI:10.1186/s12870-020-02625-8
|
| [80] |
Adhikari BN, Savory EA, Vaillancourt B, et al. Expression profiling of Cucumis sativus in response to infection by Pseudoperonospora cubensis. PLoS One, 2012, 7(4): e34954. DOI:10.1371/journal.pone.0034954
|
| [81] |
Luan Q, Chen C, Liu M, et al. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci, 2019, 279: 59-69. DOI:10.1016/j.plantsci.2018.11.002
|
| [82] |
Lai W, Zhu C, Hu Z, et al. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber. Biochem Genet, 2021, 59(4): 884-901. DOI:10.1007/s10528-021-10036-z
|
| [83] |
Perruc E, Charpenteau M, Ramirez BC, et al. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J, 2004, 38(3): 410-420. DOI:10.1111/j.1365-313X.2004.02062.x
|
| [84] |
Jing Y, Lin R. The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol, 2015, 169(1): 371-378. DOI:10.1104/pp.15.00788
|
| [85] |
Petersen K, Qiu JL, Lütje J, et al. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS One, 2010, 5(12): e14364. DOI:10.1371/journal.pone.0014364
|
| [86] |
Xie YD, Li W, Guo D, et al. The Arabidopsis gene SIGMA Factor-binding protein 1 plays a role in the salicylate-and jasmonate-mediated defence responses. Plant Cell Environ, 2010, 33(5): 828-839.
|
| [87] |
Fiil BK, Petersen M. Constitutive expression of MKS1 confers susceptibility to Botrytis cinerea infection independent of PAD3 expression. Plant Signal Behav, 2011, 6(10): 1425-1427. DOI:10.4161/psb.6.10.16759
|
| [88] |
Lai Z, Li Y, Wang F, et al. Arabidopsis Sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell, 2011, 23(10): 3824-3841. DOI:10.1105/tpc.111.090571
|
| [89] |
Wang Y, VandenLangenberg K, Wehner TC, et al. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor Appl Genet, 2016, 129(8): 1493-1505. DOI:10.1007/s00122-016-2719-x
|
| [90] |
Yoshioka Y, Sakata Y, Sugiyama M, et al. Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica, 2014, 198(2): 265-276. DOI:10.1007/s10681-014-1102-8
|
| [91] |
Vandenlangenberg KM. Studies on downy mildew resistance in cucumber (Cucumis sativus L. )[D]. Raleigh: North Carolina State University, 2015.
|
| [92] |
Li L, He H, Zou Z, et al. QTL analysis for downy mildew resistance in cucumber inbred line PI 197088. Plant Dis, 2018, 102(7): 1240-1245. DOI:10.1094/PDIS-04-17-0491-RE
|
| [93] |
Szczechura W, Staniaszek M, Klosinska U, et al. Molecular analysis of new sources of resistance to Pseudoperonospora cubensis (berk.et curt.) rostovzev in cucumber. Russ J Genet, 2015, 51(10): 1134-1140.
|
| [94] |
Win KT, Vegas J, Zhang C, et al. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor Appl Genet, 2017, 130(1): 199-211. DOI:10.1007/s00122-016-2806-z
|
| [95] |
Wang Y, VandenLangenberg K, Wen C, et al. QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by- sequencing in RIL population. Theor Appl Genet, 2018, 131(3): 597-611. DOI:10.1007/s00122-017-3022-1
|
| [96] |
Zhang K, Wang X, Zhu W, et al. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor Appl Genet, 2018, 131(10): 2229-2243. DOI:10.1007/s00122-018-3150-2
|
| [97] |
Liu XP, Lu HW, Liu PN, et al. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants- Basel, 2020, 9(12): 1659. DOI:10.3390/plants9121659
|
| [98] |
Holdsworth WL, Summers CF, Glos M, et al. Development of downy mildew-resistant cucumbers for late-season production in the northeastern United States. HortScience, 2014, 49(1): 10-17. DOI:10.21273/HORTSCI.49.1.10
|
| [99] |
翁祖信, 冯东昕, 李宝栋. 黄瓜霜霉病抗病性鉴定技术研究初报. 中国蔬菜, 1991(4): 7-9. Weng ZX, Feng DX, Li BD. Preliminary study on resistance identification technology of cucumber downy mildew. China Vegetables, 1991(4): 7-9 (in Chinese). |
| [100] |
张艳菊, 张宏宇, 秦智伟, 等. 黄瓜霜霉菌毒性及分子多态性分析. 东北农业大学学报, 2010, 41(2): 25-30. Zhang YJ, Zhang HY, Qin ZW, et al. Analysis of virulence and molecular polymorphism of Pseudoperonospora cubensis. J Northeast Agric Univ, 2010, 41(2): 25-30 (in Chinese). DOI:10.3969/j.issn.1005-9369.2010.02.006 |
| [101] |
杨柳燕, 徐永阳, 徐志红, 等. 甜瓜霜霉病抗性遗传及SRAP分子标记. 江苏农业学报, 2012, 28(5): 1200-1202. Yang LY, Xu YY, Xu ZH, et al. Inheritance of downy mildew resistance in melon and SRAP marker linked to resistant genes. Jiangsu J Agric Sci, 2012, 28(5): 1200-1202 (in Chinese). |
| [102] |
刘通. 不同地区黄瓜霜霉病菌毒力的差异. 北方园艺, 2011(14): 141-144. Liu T. Study on virulence differences of cucumber downy mildew in different areas. North Hortic, 2011(14): 141-144 (in Chinese). |
| [103] |
Nie JT, Wang H, Zhang WL, et al. Characterization of lncRNAs and mRNAs involved in powdery mildew resistance in cucumber. Phytopathology, 2021, 111(9): 1613-1624. DOI:10.1094/PHYTO-11-20-0521-R
|
| [104] |
黄金存, 叶冰莹, 许玉芬, 等. 转录因子WRKY和NPR1在系统获得抗性信号转导中的相互作用机制. 生物技术通讯, 2007, 18(6): 992-994. Huang JC, Ye BY, Xu YF, et al. The interaction mechanism between transcription factor WRKY and NPR1 in systemic acquired resistance signal transduction. Lett Biotechnol, 2007, 18(6): 992-994 (in Chinese). DOI:10.3969/j.issn.1009-0002.2007.06.031 |
 2022, Vol. 38
2022, Vol. 38