中国科学院微生物研究所、中国微生物学会主办
文章信息
- 刘欢, 陈旺, 谭森文, 梁思雨, 杨晨曦, 张千
- LIU Huan, CHEN Wang, TAN Senwen, LIANG Siyu, YANG Chenxi, ZHANG Qian
- 不动杆菌Acinetobacter sp. TAC-1利用聚(3-羟基丁酸酯-co-3-羟基戊酸酯)的碳代谢机理
- An examination of the carbon metabolic pathways in Acinetobacter sp. TAC-1 in the context of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) utilization
- 生物工程学报, 2023, 39(11): 4663-4681
- Chinese Journal of Biotechnology, 2023, 39(11): 4663-4681
- 10.13345/j.cjb.230120
-
文章历史
- Received: February 19, 2023
- Accepted: July 4, 2023
- Published: July 18, 2023
传统生物脱氮技术具有适用范围广、处理效率高和运行成本低等优势,是城镇污水处理中使用最广泛的脱氮技术。近几年发布的中国环境公报显示,湖泊富营养化、河道黑臭和地下水污染的形势仍然十分严峻。为此,《城镇污水处理厂污染物排放标准》(GB18918—2002)[1]中对氮的排放提出了更高的要求[总氮(total nitrogen, TN)的排放限值从20 mg/L降到15 mg/L],而传统生物脱氮技术出现了低温硝化和低碳源反硝化两大瓶颈问题。一是低温对硝化菌的比增长速率和活性都有强烈抑制。当水温在10 ℃左右时,每降低1 ℃,硝化活性会降低50%[2]。因此,排放标准里指出,当温度低于12 ℃时,一级A标对氨氮排放的浓度限值从5 mg/L放宽到8 mg/L。二是低碳源导致反硝化过程电子供体缺乏。当BOD/TN < 4时,反硝化过程会受到明显抑制[3]。目前一级A中TN 15 mg/L的限值对大多数污水处理厂来说仍然面临巨大压力。因此,寻求新的生物脱氮技术,成为解决城镇污水处理厂提标改造的关键。
异养硝化-好氧反硝化(heterotrophic nitrification-aerobic denitrification, HN-AD)菌属在低温低碳条件下的脱氮潜能逐渐被挖掘。HN-AD菌属不仅可以在低温条件下(2−15 ℃)保持较高的脱氮速率[4-11],还可以在低碳源条件下通过异养硝化和好氧反硝化实现氨氮和总氮的有效去除[12-15],因此在低温低碳废水的处理中具备重大潜力和优势。然而多数HN-AD菌分离自水处理系统,它们在原有水处理系统中并未形成优势菌属,为了拓展HN-AD菌的应用,研究者尝试通过固定化技术来实现HN-AD菌在水处理系统里的富集[16-21],尽管采用的固定化技术有一定富集效果,但在废水处理系统中由于碳源竞争能力较弱,导致HN-AD菌容易失去优势地位。在固定化材料中,固体碳源固定体系可以有效固定反硝化功能微生物进行高效脱氮成为研究热点,其中研究最广泛的固体碳源类型主要有聚(3-羟基丁酸酯-co-3-羟基戊酸酯) [poly (3-hydroxybutyrate-co-3-hydroxyvalerate), PHBV]、聚己内酯(polycaprolactone, PCL)、聚丁二酸丁二醇酯(polybutylene succinate, PBS)和聚乳酸(polylactic acid, PLA)等[22]。这些固体碳源降解后可以分解为羟基丁酸、羟基乙酸、乙酸、丁二酸和羟基丙酸等小分子有机酸[23-24]。目前固体碳源微生物降解的研究主要聚焦于土壤环境中微生物对固体碳源薄膜的降解,降解机制也主要集中于降解酶的酶活、性能验证以及降解产物的研究上[25],对于降解酶类型以及降解产物的代谢则鲜有报道。关于HN-AD菌利用固体碳源PHBV的最新研究也多关注其异养硝化-好氧反硝化作用[26-29],对碳代谢机制的阐述鲜有报道。
本研究采用HN-AD菌不动杆菌(Acinetobacter sp.) TAC-1与固体碳源PHBV进行实验。以SOA为对照,采用基因测序、非靶向代谢组学等分析方法,研究降解酶类型和对应碳代谢通路以及降解产物差异,以期阐释TAC-1菌利用固体碳源PHBV的碳代谢机理。本研究有望为基于HN-AD和固体碳源的脱氮新工艺的开发和应用提供理论指导。
1 材料与方法 1.1 TAC-1菌株来源及培养本研究使用菌株为一株具有低温高效脱氮功能的不动杆菌TAC-1,从重庆市某养猪场沼液中分离得到,详细的分离过程见团队前期的研究报道[30]。将TAC-1菌株置于100 mL灭菌培养基,培养基组成:0.547 g CH3COONa、0.189 g (NH4)2SO4、5 mL微量元素,PH值7.0−7.2;0.2 MPa、120 ℃下灭菌30 min;其中微量元素组成为(g/L):MgSO4·7H2O 2.0,MnSO4·H2O 0.1,CaCl2 1.5,FeSO4·7H2O 0.1,K2HPO4 5.0。在恒温摇床(30 ℃、150 r/min)中培养24 h。
1.2 DNA提取及全基因测序将1.1节所述方式培养的TAC-1菌离心收集。按照上海美吉生物医药科技有限公司提供的说明书,使用MobioPowerSoil® DNA分离试剂盒提取DNA样本。随后,将提取的DNA样本送至上海美吉生物医药科技有限公司进行全基因测序。所有测序结果在上海美吉生物云平台(www.majorbio.com)上进行分析。
1.3 TAC-1菌和固体碳源实验实验组:在500 mL已灭菌的锥形瓶中依次加入30 mL TAC-1菌、300 mL灭菌自来水、30 g固体碳源(PHBV)。
对照组:在500 mL已灭菌的锥形瓶中依次加入30 mL TAC-1菌30、300 mL灭菌自来水、3.48 g无水乙酸钠(sodium acetate, SOA) (选择依据来自前期TAC-1菌碳源优选实验[30]),初始化学需氧量(chemical oxygen demand, COD)值与30 g PHBV相同,均为1 000 mg/L。
实验在30 ℃、150 r/min的恒温摇床中进行。每12 h在无菌操作台取一次样品,取样后立即测定菌液浓度(OD600)和总有机碳(total organic carbon, TOC)浓度。所有实验测试均为3次,数据取平均值。
1.4 总有机碳(total organic carbon, TOC)与菌液浓度(OD600)的检测方法TOC采用燃烧氧化-非色散红外检测法测定[31],OD600采用紫外分光光度法测定。
1.5 非靶向代谢组学分析锥形瓶实验运行5 d后,从SOA和PHBV组中采集生物样品(每组采集6个样本,每个样本重量为0.15 g)。将样品迅速冷冻于干冰中并送至上海中科新生命生物科技有限公司的代谢组分析平台,进行非靶向代谢组学检测分析。
2 结果与分析 2.1 TAC-1菌的基本基因组信息利用Circos软件绘制基因组圈图,用于可视化基因组的结构和关系,如图 1所示,TAC-1菌包含1个染色体和3个质粒。
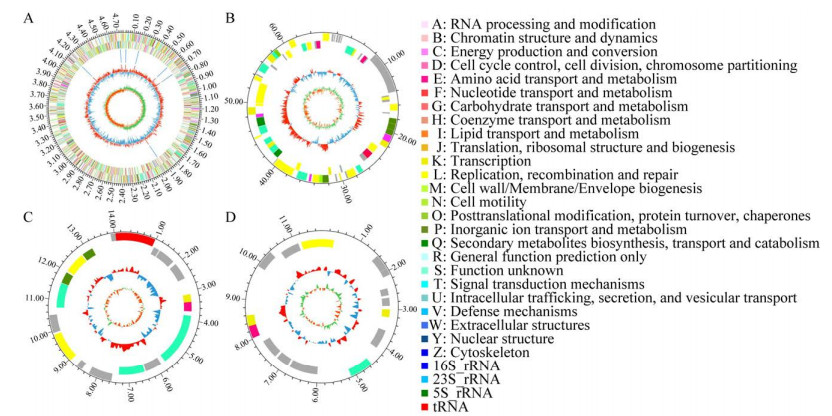
|
| 图 1 TAC-1细菌基因组圈图 Fig. 1 The TAC-1 bacterial genome ring diagram. A:染色体. B−D:3个质粒. 不同的颜色表示编码序列不同的COG的功能分类 A: Chromosomes. B–D: Plasmids. Distinct colors signify the functional classification of COG with different coding sequences. |
| |
如表 1所示,预测TAC-1基因组共编码4 660个基因,基因组总长度为4 862 297 bp,编码基因总长度为4 103 118 bp,占基因组的84.39%,编码基因平均长度为888.50 bp。共查到81个tRNA和21个rRNA基因。GC含量为38.33%。
| Project | Numerical value |
| Genome size (bp) | 4 862 297 |
| Gene total len (bp) | 4 103 118 |
| Gene/Genome (%) | 84.39 |
| CDS No. | 4 660 |
| Gene average len (bp) | 888.50 |
| GC content (%) | 38.33 |
| tRNA No. | 81 |
| rRNA No. | 21 |
通过与KEGG数据库比对,有2 174个基因在KEGG数据库中被匹配,包括39个KEGG通路(图 2)。在level 1层级分类的6个通路中,代谢(metabolism)为最高水平,图中共检测到1 909个(87.81%)与代谢相关的基因,其次是环境信息处理(environmental information processing) 230个(10.58%)、遗传信息加工(genetic information processing) 184个(8.46%)、人类疾病(human diseases) 128个(5.88%)、细胞过程(cellular processes) 127个(5.84%)、有机体系统(organismal systems) 56个(2.58%)。而在代谢功能作用中,碳水化合物代谢的基因为156个(占比8.17%)。因此,TAC-1菌株存在与重要代谢过程相对应的基因。
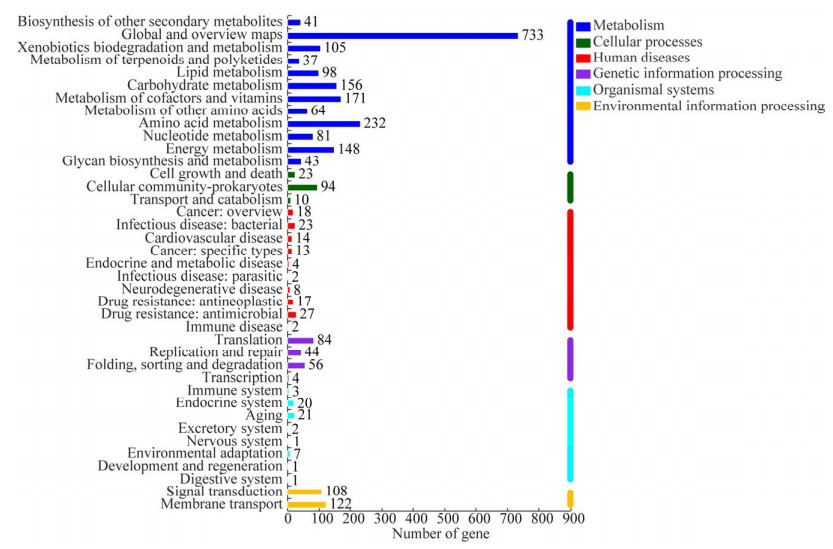
|
| 图 2 TAC-1基因代谢通路 Fig. 2 The metabolic pathways of genes in the Acinetobacter sp. TAC-1 strain as characterized through genomics analysis. 图中的6种颜色分别代表 6个主要的通路水平,深蓝色表示代谢;深绿色表示细胞过程;红色表示人类疾病;紫色表示遗传信息过程;青色表示有机系统;橙色表示环境信息过程 The six colors in the figure respectively represent the six main pathway levels, with dark blue representing metabolism, dark green representing cellular processes, red representing human diseases, purple representing genetic information processes, cyan representing organic systems, and orange representing environmental information processes. |
| |
对图 2的碳水化合物部分相关基因进行KEGG功能注释分析,得到如表 2所示的TAC-1菌株的碳代谢基因与对应编码的酶。TAC-1菌株编码表达碳代谢相关酶的基因如pckA (编码磷酸烯醇丙酮酸羧激酶)、gltA (编码柠檬酸合成酶)、acnA (编码乌头酸水合酶)、icd (编码异柠檬酸脱氢酶)、sucAB (编码2-氧戊二酸脱氢酶)、sucD (编码琥珀酰辅酶A合成酶)、aarC (编码乙酸辅酶A转移酶)、sdhA (编码琥珀酸脱氢酶)、fumA (编码延胡索酸水合酶)、mqo (编码苹果酸脱氢酶)、mdh (编码苹果酸脱氢酶)与发生在细菌内的TCA循环反应有关[32]。刘钊远[33]通过重组柠檬酸合成酶、异柠檬酸脱氢酶和α-酮戊二酸脱氢酶过表达基因工程菌株,与原始菌株进行对比发酵验证,发现游离表达icd基因的工程菌杆菌肽产量比原始菌株高出11.5%,同时编码基因gltA的转录水平比原始菌株提高了3.4倍。由此推测,在TCA循环中,起到调节作用的编码基因为gltA、icd。
| Gene ID | KEGG name | KO description |
| Gene 3461 | pckA | Phosphoenolpyruvate carboxykinase (GTP) |
| Gene 1134 | gltA | Citrate synthase |
| Gene 3980 | acnA | Aconitate hydratase |
| Gene 1274 | icd | Isocitrate dehydrogenase |
| Gene 1139 | sucA | 2-oxoglutarate dehydrogenase E1 component |
| Gene 1140 | sucB | 2-oxoglutarate dehydrogenase E2 component |
| Gene 1143 | sucD | Succinyl-CoA synthetase alpha subunit |
| Gene 0266 | aarC | Acetate CoA-transferase |
| Gene 1137 | sdhA | Succinate dehydrogenase |
| Gene 3995 | fumA | Fumarate hydratase, class Ⅰ |
| Gene 2554 | mqo | Malate dehydrogenase (quinone) |
| Gene 4044 | mdh | Malate dehydrogenase |
| Gene 1776 | acs | Acetyl-CoA synthetase |
蛋白质直系同源簇(cluster of orthologous groups of proteins, COG)数据库的基因注释统计结果如图 3所示。注释结果显示TAC-1基因的COG功能分类共有22个,共注释到3 602个基因,其中33.29%的基因没有功能特征。第二大和第三大功能分类分别是“转录”和“氨基酸转运与代谢”,它们分别注释268个和244个基因。其次第三大和第四大功能分类分别是“无机离子运输和代谢”和“能源生产和转换”,它们分别注释235个和217个基因。COG注释的TAC-1菌株部分碳代谢基因如表 3所示。
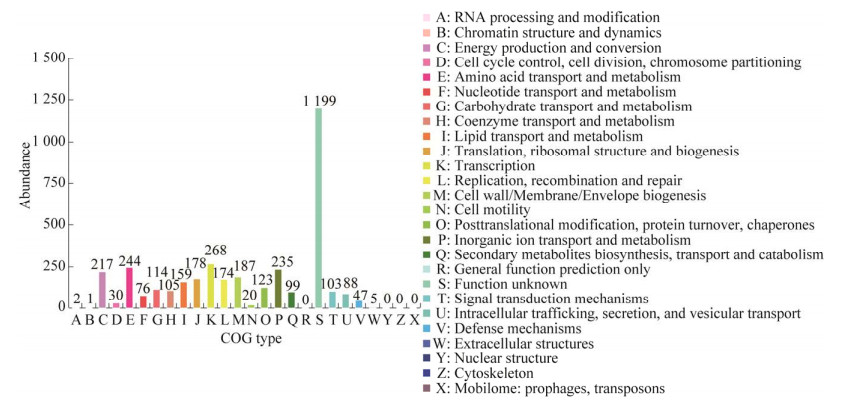
|
| 图 3 COG注释 Fig. 3 COG annotations. 横坐标代表不同COG类型,纵坐标代表基因个数. 具体每种COG类型的功能描述请见右侧图例 The x-axis represents different COG types, and the y-axis represents the number of genes. Detailed functional descriptions for each COG type can be found in the legend on the right side. |
| |
| Gene ID | COG description | COG type description | COG category |
| Gene 4563 | Phosphoenolpyruvate carboxylase activity | Energy production and conversion | Metabolism |
| Gene 0266 | Acetyl-CoA hydrolase | Energy production and conversion | Metabolism |
| Gene 1134 | Citrate synthase | Energy production and conversion | Metabolism |
| Gene 3709 | Citrate transporter | Energy production and conversion | Metabolism |
| Gene 2405 | Fumarate hydratase class Ⅱ | Energy production and conversion | Metabolism |
| Gene 3995 | Fumarate | Energy production and conversion | Metabolism |
| Gene 1274 | Isocitrate dehydrogenase | Energy production and conversion | Metabolism |
| Gene 3706 | Isocitrate lyase | Energy production and conversion | Metabolism |
| Gene 0142 | Malic enzyme | Energy production and conversion | Metabolism |
| Gene 2554 | Malate dehydrogenase (quinone) | Energy production and conversion | Metabolism |
| Gene 0116 | NADH flavin oxidoreductase | Energy production and conversion | Metabolism |
| Gene 1244 | NADH dehydrogenase | Energy production and conversion | Metabolism |
| Gene 1137 | Succinate dehydrogenase | Energy production and conversion | Metabolism |
| Gene 2272 | Citrate lyase | Carbohydrate transport and metabolism | Metabolism |
| Gene 2881 | (ABC) transporter | Carbohydrate transport and metabolism | Metabolism |
| Gene 2887 | Carbohydrate kinase | Carbohydrate transport and metabolism | Metabolism |
基因本体论(gene ontology, GO)数据库的基因注释统计结果如图 4所示,共注释到3 026个基因(占总编码基因数的64.93%)。其中分子功能(molecular function, MF)丰度最高(基因数目2 395,79.15%),其次是细胞组分(cellular component, CC) (基因数目1 454,48.05%)和生物过程(biological process, BP) (基因数目1 351,44.65%)。而在分子功能中主要富集到ATP结合相关基因。GO注释的TAC-1菌株部分碳代谢相关功能基因数目如表 4所示。
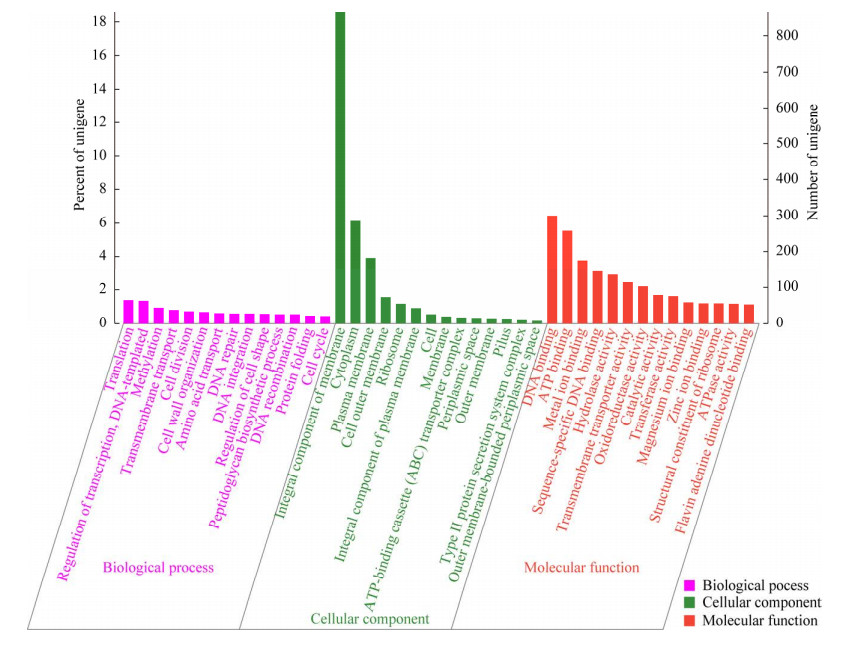
|
| 图 4 GO注释 Fig. 4 GO annotations. 横坐标代表GO的三大分支(即生物过程、细胞组分、分子功能)以及更进一步的leve 2分类;纵坐标代表基因所占的相对比例 The x-axis represents the three major branches of gene ontology (GO): Biological processes, cellular components, and molecular functions, along with further level 2 classifications. The y-axis represents the relative proportion of genes. |
| |
| Category | GO description | Gene No. |
| Molecular function | ATP binding | 258 |
| Molecular function | NADH dehydrogenase (ubiquinone) activity | 11 |
| Molecular function | Acetyl-CoA carboxylase activity | 4 |
| Molecular function | Acetyl-CoA C-acetyltransferase activity | 4 |
| Molecular function | Carbohydrate derivative binding | 3 |
| Molecular function | Gluconate transmembrane transporter activity | 3 |
| Molecular function | Fumarate hydratase activity | 2 |
| Molecular function | Isocitrate dehydrogenase (NADP+) activity | 2 |
| Molecular function | Acetate-CoA ligase activity | 1 |
| Molecular function | Malate dehydrogenase (decarboxylating) (NADP+) activity | 1 |
| Molecular function | Isocitrate lyase activity | 1 |
| Molecular function | Phosphoenolpyruvate carboxykinase (GTP) activity | 1 |
| Molecular function | Succinate dehydrogenase activity | 1 |
| Molecular function | Glucose-6-phosphate isomerase activity | 1 |
| Molecular function | Acetyl-CoA hydrolase activity | 1 |
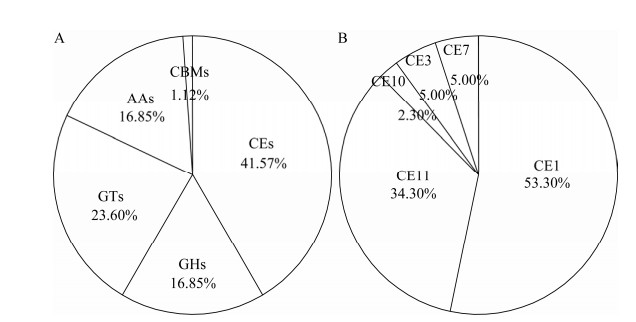
碳水化合物活性酶(carbohydrate active enzymes, CAZyme)包括多个不同的酶家族,其中包括糖苷水解酶(glycoside hydrolases, GHs)、糖基转移酶(glycosyltransferases, GTs)、多糖裂合酶(polysaccharide lyases, PLs)、碳水化合物酯酶(carbohydrate esterases, CEs)、碳水化合物结合模块(carbohydrate-binding modules, CBMs)和辅助氧化还原酶(auxiliary activities, AAs)。这些酶在降解复杂碳水化合物方面发挥关键作用,其中GHs通过水解糖苷键分解多糖[34-35],GTs参与糖类物质的合成[36],CEs降解多糖酯基,促进其他酶的水解作用[37],PLs通过复杂机制破坏糖苷键[38],CBMs则通过将酶固定到底物表面提高水解效率[39-40],AAs可参与木质素修饰,打开植物生物质抗降解屏障,加速底物水解[41]。细菌产生的碳水化合物酶协同作用可将复杂的碳水化合物降解为简单糖类。
菌株TAC-1中CAZyme基因表达结果见图 5A,分析显示GHs、CEs、GTs、AAs和CBMs基因表达比例分别为16.85%、41.57%、23.60%、16.85%和1.12%。图 5B表明,CEs基因表达最为丰富,其中主要种类包括CE11 (葡萄糖脱乙酰基酶)、CE1 (羧酸酯酶、木聚糖酯酶、阿魏酰酯酶、甘肽水解酶、酰基转移酶、芳基酯酶、胆碱酯酶、甾醇酯酶和羧基酯酶)[42]。
|
| 图 5 碳水化合物活性酶的表达量(A)和碳水化合物酯酶的表达量(B) Fig. 5 Expression of carbohydrate active enzymes (A) and carbohydrate esterases (B). 饼图的面积表示该分类中基因所占的相对比例 The area of the pie chart indicates the relative proportion of gene expression in that classification. |
| |
上述分析表明,碳水化合物酶如CEs的高量表达,可能是TAC-1菌降解PHBV的关键。
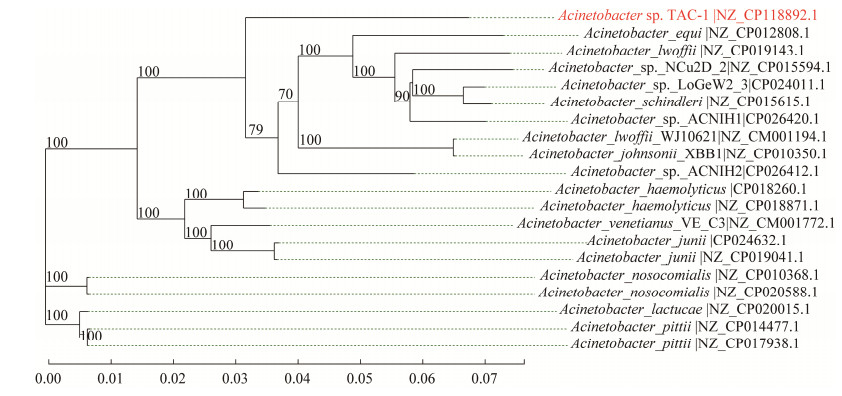
2.5 系统发育树使用31个持家基因(dnaG、frr、infC、nusA、pgk、pyrG、rplA、rplB、rplC、rplD、rplE、rplF、rplK、rplL、rplM、rplN、rplP、rplS、rplT、rpmA、rpoB、rpsB、rpsC、rpsE、rpsI、rpsJ、rpsK、rpsM、rpsS、smpB和tsf)与本地数据库进行比对,并从中选择与目标菌株在种属水平上最为相似的19株菌进行进化分析。
如图 6所示,TAC-1菌株与lwoffii_WJ10621亲缘关系较近,相似度高达92.8%。
|
| 图 6 基于持家基因的TAC-1菌株系统发育树 Fig. 6 The phylogenetic tree of TAC-1 strain based on housekeeping genes. |
| |
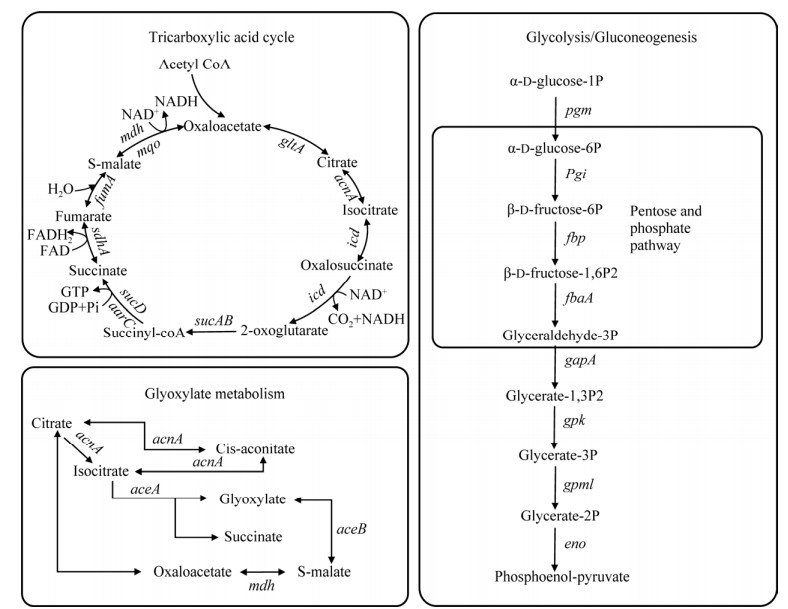
将上述分析的酶及编码基因结合KEGG数据库、COG数据库和GO数据库的功能注释以及KEGG通路的预测分析,得到如图 7所示TAC-1菌存在的碳代谢路径[32]。结果表明,TAC-1菌存在糖醇解、磷酸戊糖途径、三羧酸循环和乙醛酸代谢等碳代谢路径。然而,TAC-1菌利用PHBV的中间产物还未可知,仍需进一步分析。
|
| 图 7 TAC-1菌存在的部分碳代谢路径(包括糖醇解、磷酸戊糖途径、三羧酸循环和乙醛酸代谢) Fig. 7 Partial carbon metabolism pathways of TAC-1 bacteria, including glycolysis, pentose phosphate pathway, tricarboxylic acid cycle and glyoxylic acid metabolism. |
| |
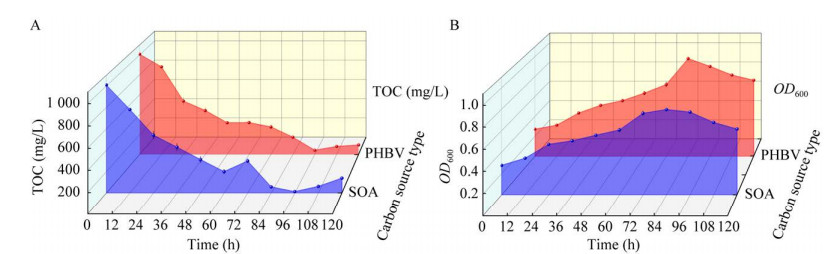
如图 8所示,实验组使用PHBV和对照组使用SOA的TOC和OD600均呈现出类似的趋势。初始TOC为1 000 mg/L,之后TOC快速下降,同时OD600呈上升趋势。这表明TAC-1菌能够有效利用PHBV和SOA,并且在利用程度和生长趋势上表现出相似的结果。然而,当TOC浓度降至较低水平(< 200 mg/L)时,TAC-1菌的生长受到限制,甚至导致OD600的下降。
|
| 图 8 TAC-1菌利用不同碳源的TOC (A)与OD600 (B)变化情况 Fig. 8 TOC (A) and OD600 (B) changes of TAC-1 bacteria using different carbon sources. 垂直坐标表示TOC浓度(A)和OD600 (B),水平坐标表示反应时间,水平向里的坐标表示碳源类型 The vertical axis represents TOC concentration (A) and OD600 (B), while the horizontal axis represents reaction time. The inward horizontal coordinate represents the carbon source type. |
| |
上述结果表明,反应期间细菌数量增长速率不同或碳源利用程度不同不是造成后续分析结果差异的因素,碳源类型的不同才是引起后续分析的差异的因素。
2.7.2 不同碳源下差异性代谢物分析不同类型碳源提取的两组样本和QC样本正、负离子模式下PCA得分图和火山图如图 9所示,结果显示在正离子和负离子模式下,SOA和PHBV组间的代谢产物分离情况良好。本次在正、负离子模式下分别检测到7 356个离子峰,图中显示在正、负离子模式下,以SOA为对照组,PHBV因其更大的表面积附着更多的菌导致丰度显著上调的代谢物数量均高于丰度显著下降的代谢物数量[43]。
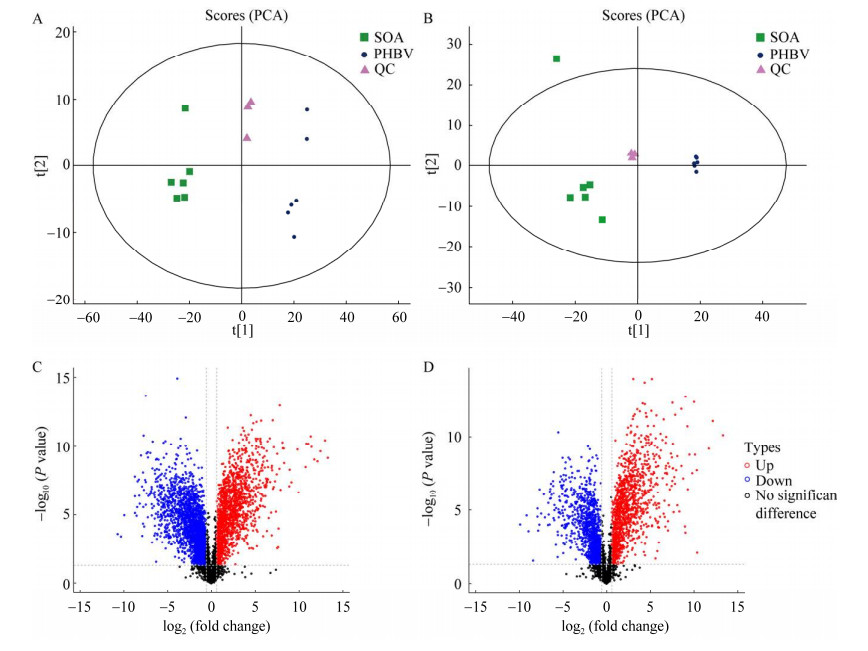
|
| 图 9 正负离子模式下的PCA得分图和代谢物丰度差异火山图 Fig. 9 PCA score plot and volcano plots. A、C:正离子模式. B、D:负离子模式. (A−B)中每个点代表一个样本,其位置表示在降维空间中的得分. 不同颜色或标记可以表示不同的组或类别,轴标签说明了各个主成分的贡献程度. (C−D)中红色表示差异代谢物中存在显著上调的变化(FC > 1.5,P < 0.05),蓝色表示差异代谢物中存在显著下调的变化(FC < 0.67,P < 0.05),黑色表示差异代谢物没有显著性变化 A, C: Positive mode. B, D: Negative mode. Each point in (A−B) represents a sample, and its position reflects the score in the reduced dimensional space. Different colors or markers can be used to represent different groups or categories, while the axis labels indicate the contribution levels of each principal component. In (C−D), the red color indicates significant upregulation of differential metabolites (FC > 1.5, P < 0.05), the blue color represents significant downregulation of differential metabolites (FC < 0.67, P < 0.05), and the black color signifies no significant change in differential metabolites. |
| |
经过KEGG富集分析,确定了具有显著差异的前20条代谢途径(P < 0.05) (图 10)。结果表明PHBV对TAC-1菌的代谢途径,尤其是在氨酰基-tRNA合成、脂肪酸合成和不饱和脂肪酸合成、ABC转运途径以及氰基氨基酸代谢等通路,产生了明显影响。
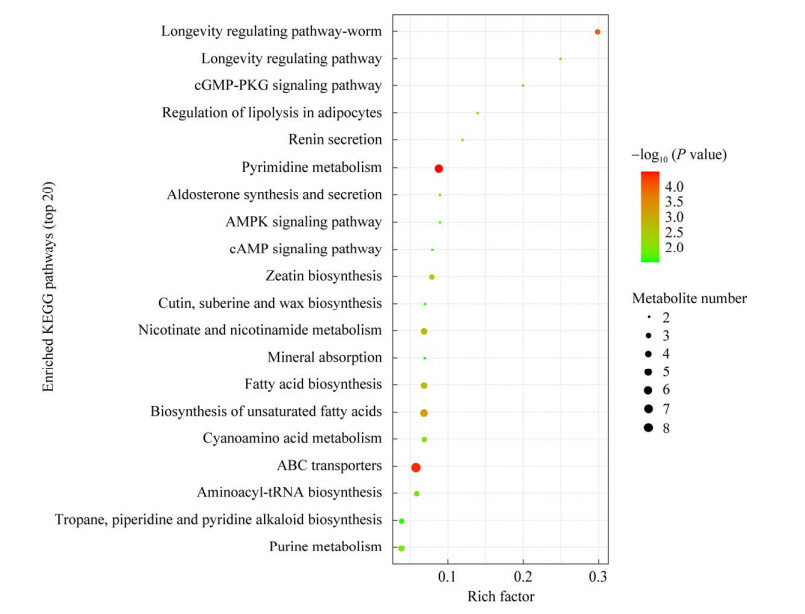
|
| 图 10 KEGG富集通路图 Fig. 10 KEGG enrichment pathway plot. 圆圈大小表示该通路的代谢物数量,颜色代表富集的P值 The size of the circles represents the number of metabolites in that pathway. The color represents the enrichment P value. |
| |
通过进一步分析在通路活性上显示明显差异的代谢途径,利用差异丰度得分来评估代谢的变化情况,并捕捉特定途径中所有代谢物的平均和整体变化。根据Pathway_hierarchy1的分类归属,将这些途径划分为环境信息处理、遗传信息处理、代谢机制和有机体系统等4大类别。在本研究中,共发现了24条显著差异的代谢通路,其中有20条上调和4条下调,结果见图 11。其中微生物新陈代谢(如戊糖磷酸代谢)、遗传信息处理(如氨酰基-tRNA的生物合成)以及有机体系统(如脂肪细胞脂肪分解的调节)等相关代谢通路的活性在添加固体碳源PHBV的作用下显著上调,说明在固体碳源PHBV的系统内TAC-1菌群细胞的生长代谢繁殖及遗传表达起着正向作用。
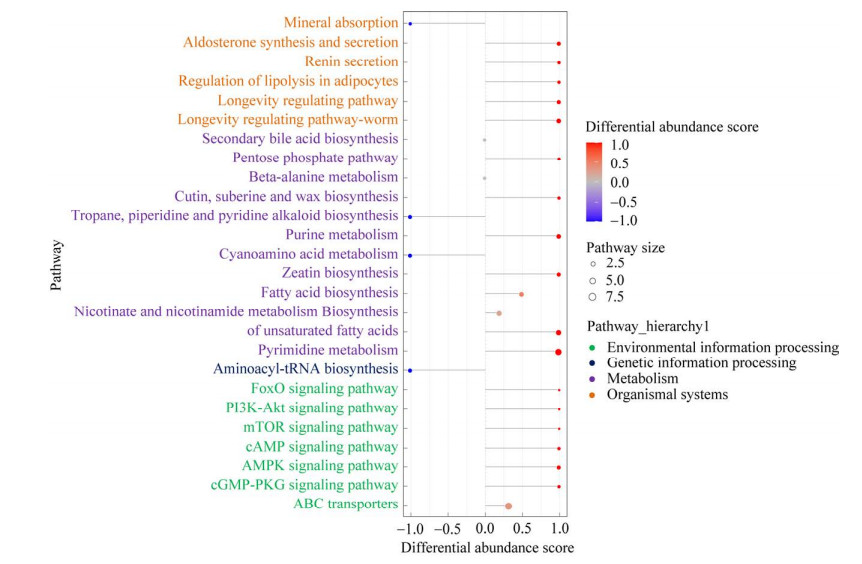
|
| 图 11 差异代谢通路的差异丰度得分图 Fig. 11 Analysis of different abundance of metabolic pathways based on PHBV_vs._SOA. 线段的长度表示差异丰度得分的绝对值,圆点大小表示该通路中代谢物的数目;红色的深度表示该通路整体表达情况上调程度,蓝色的深度表示该通路整体表达情况下调程度 The length of the line segment indicates the absolute value of the differential abundance score, and the size of the dots indicates the number of metabolites in the pathway; The depth of red color represents the overall upregulation level of the pathway, while the depth of blue color represents the overall downregulation level of the pathway. |
| |
如图 11所示,对于FoxO信号通路,FoxO转录因子家族对细胞生理事件中的基因表达起着调控作用,包括凋亡、细胞周期控制、葡萄糖代谢、抗氧化应激和长寿等重要过程[44]。在环境信息处理中,cGMP-PKG和PI3K-Akt信号通路得到上调。这2个信号通路与细胞的生长增殖密切相关,其中cGMP-PKG信号通路主要推动细胞的生长增殖过程,而作为PI3K信号通路下游的主要应激酶,Akt参与细胞的生长、分化和损伤保护等重要过程[45-46]。此外,ABC转运蛋白在微生物抗逆性中发挥重要作用,负责转运无机盐离子、糖和氨基酸等关键物质[47]。
上述信号通路活性在实验组上调,说明相较SOA,固体碳源PHBV可强化TAC-1菌群的细胞生长与代谢。
2.7.4 TAC-1菌利用PHBV的碳代谢机制功能微生物可以通过糖酵解途径(EMP)、磷酸戊糖途径(PPP)生成乙酰辅酶A进入三羧酸循环[48-50],从而持续产生电子供体NADH和释放大量ATP供能[51]。ATP是代谢反应和通路最重要的能量来源,在菌株的生长和目标代谢产物的合成中起着至关重要的作用[52]。同时,在代谢途径中共享的辅酶因子ATP/ADP、NADH/NAD和acetyl-CoA/辅酶A在代谢通量分布和速率中也起着重要作用[53-57]。在实验组PHBV中,能量代谢的中间产物NAD+、NADH、ADP和ATP相较于对照组SOA分别提高了5、2.73、2.57、6.18倍(基于非靶向代谢结果报告计算得出),说明添加固体碳源PHBV能够提升菌群能量和代谢水平。检测到PPP路径中代谢产物葡萄糖酸盐的上调说明PHBV碳源大分子被分解成小分子葡萄糖酸盐,进而被TAC-1菌在细胞内利用。Acetyl-CoA作为碳源和TCA循环之间的重要桥梁,其上调的代谢结果加快了碳水化合物代谢,说明PHBV更能有效地被TAC-1菌株利用。与之不同,乙酸钠可直接通过乙酸代谢产生乙酰辅酶A (acetyl-CoA)进入TCA循环[54]。如图 12所示,在TCA循环的第一步,乙酰辅酶A中的乙酰基转移到草酰乙酸,生成柠檬酸。接下来的2个氧化脱羧步骤中,产生2-酮戊二酸和琥珀酰辅酶A,同时放出二氧化碳和NADH。琥珀酰辅酶A与琥珀酸反应时,产生GTP,用于生成ATP。琥珀酸脱氢酶将琥珀酸转化为富马酸,产生FADH、苹果酸和草酰乙酸,形成TCA循环的下一轮。同时,由EMP途径产生的丙酮酸可以进入TCA循环被氧化,或被羧化成草酰乙酸。
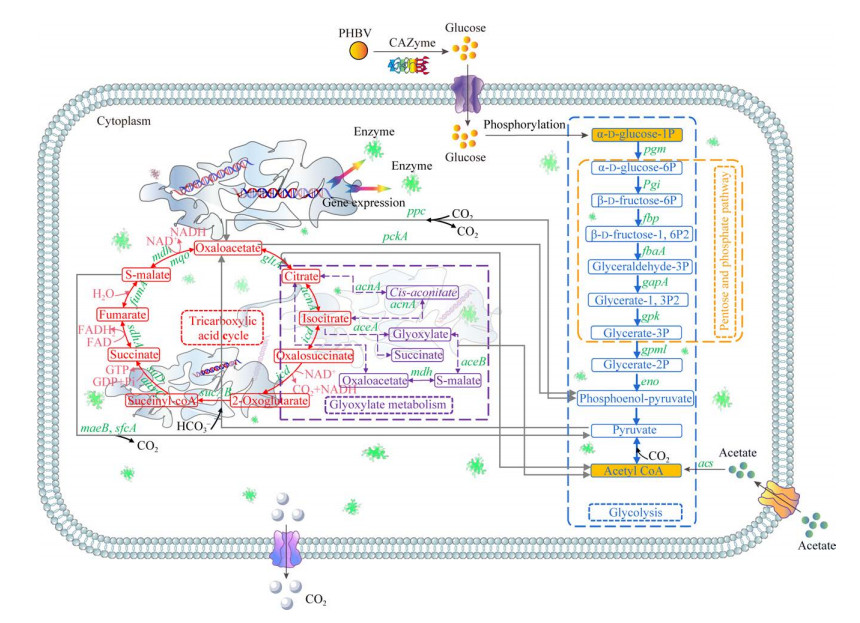
|
| 图 12 TAC-1菌在PHBV作用下的碳代谢机制 Fig. 12 The metabolic pathway of Acinetobacter sp. TAC-1 in the presence of PHBV as a solid carbon source. 图中蓝色虚线表示EMP;黄色虚线表示PPP;红色线部分为TCA循环;紫色虚线部分表示乙醛酸代谢 The blue dotted line signifies the glycolytic pathway (EMP), the yellow dotted line represents the pentose phosphate pathway (PPP), the red line represents the tricarboxylic acid (TCA) cycle, and the purple dotted line represents the metabolism of glyoxylic acid. |
| |
本研究采用基因组测序对TAC-1菌株进行分析,TAC-1菌株在基因水平上存在编码碳水化合物酶的关键基因,包括gltA、icd、sucAB、acs和pckA等。通过KEGG数据库注释,进一步证实了TAC-1菌株存在着碳水化合物代谢通路,包括EMP途径、PPP途径、乙醛酸代谢和TCA循环。结合TAC-1在利用不同碳源(PHBV与SOA)时的代谢物差异分析,结果显示,与SOA组相比,PHBV组的能量代谢中间产物NAD+、NADH、ADP和ATP分别提高了5、2.73、2.57、6.18倍。此外,葡萄糖酸盐和乙酰辅酶A的表达也得到了上调。以上结果进一步证实了TAC-1菌株利用PHBV的途径为:PHBV (通过PPP途径)→葡萄糖酸盐(通过EMP途径)→acetyl-CoA (进入TCA循环)→CO2+H2O (产生电子供体并释放能量)。而对于SOA的利用途径为:SOA (通过乙酸代谢)→acetyl-CoA (进入TCA循环)→CO2+H2O (产生电子供体并释放能量)。
| [1] |
GB 18918—2002. 城镇污水处理厂污染物排放标准[S]. 北京: 中国标准出版社, 2002. GB 18918—2002. Pollutant emission standards for urban sewage treatment plants[S]. Beijing: China Standard Press, 2002 (in Chinese). |
| [2] |
HEAD MA, OLESZKIEWICZ JA. Bioaugmentation for nitrification at cold temperatures[J]. Water Research, 2004, 38(3): 523-530. DOI:10.1016/j.watres.2003.11.003
|
| [3] |
HER JJ, HUANG JS. Influences of carbon source and C/N ratio on nitrate/nitrite denitrification and carbon breakthrough[J]. Bioresource Technology, 1995, 54(1): 45-51. DOI:10.1016/0960-8524(95)00113-1
|
| [4] |
YAO S, NI J, CHEN Q, BORTHWICK AGL. Enrichment and characterization of a bacteria consortium capable of heterotrophic nitrification and aerobic denitrification at low temperature[J]. Bioresource Technology, 2013, 127: 151-157. DOI:10.1016/j.biortech.2012.09.098
|
| [5] |
YE Q, LI KL, LI ZL, XU Y, HE TX, TANG WH, XIANG SD. Heterotrophic nitrification-aerobic denitrification performance of strain Y-12 under low temperature and high concentration of inorganic nitrogen conditions[J]. Water, 2017, 9(11): 835. DOI:10.3390/w9110835
|
| [6] |
QU D, WANG C, WANG YF, ZHOU R, REN HJ. Heterotrophic nitrification and aerobic denitrification by a novel groundwater origin cold-adapted bacterium at low temperatures[J]. RSC Advances, 2015, 5(7): 5149-5157. DOI:10.1039/C4RA13141J
|
| [7] |
ZHANG D, LI W, HUANG X, QIN W, LIU M. Removal of ammonium in surface water at low temperature by a newly isolated Microbacterium sp. strain SFA13[J]. Bioresource Technology, 2013, 137: 147-152. DOI:10.1016/j.biortech.2013.03.094
|
| [8] |
冯叶. 低温冲击对异养硝化-好氧反硝化菌HN-02的影响研究[D]. 四川: 西南交通大学硕士学位论文, 2014. FENG Y. Investigation into the effect of low temperature shock on the performance of nitrifying-aerobic denitrifying bacterium HN-02 in a bioprocess system[D]. Sichuan: Master's Thesis of Southwest Jiaotong University, 2014 (in Chinese). |
| [9] |
HUANG XF, LI WG, ZHANG DY, QIN W. Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification-aerobic denitrification at low temperature[J]. Bioresource Technology, 2013, 146: 44-50. DOI:10.1016/j.biortech.2013.07.046
|
| [10] |
XU Y, HE TX, LI ZL, YE Q, CHEN YL, XIE EY, ZHANG X. Nitrogen removal characteristics of Pseudomonas putida Y-9 capable of heterotrophic nitrification and aerobic denitrification at low temperature[J]. BioMed Research International, 2017, 2017, 1-7.
|
| [11] |
何腾霞, 倪九派, 李振轮, 孙权, 冶青, 徐义. 1株Arthrobacter arilaitensis菌的耐冷异养硝化和好氧反硝化作用[J]. 环境科学, 2016, 37(3): 1082-1088. HE TX, NI JP, LI ZL, SUN Q, YE Q, XU Y. Heterotrophic nitrification and aerobic denitrification of the hypothermia aerobic denitrification bacterium: Arthrobacter arilaitensis[J]. Environmental Science, 2016, 37(3): 1082-1088 (in Chinese). |
| [12] |
ZHENG ZJ, ZHANG DY, LI WG, QIN W, HUANG XF, LV LY. Substrates removal and growth kinetic characteristics of a heterotrophic nitrifying-aerobic denitrifying bacterium, Acinetobacter harbinensis HITLi7T at 2 ℃[J]. Bioresource Technology, 2018, 259: 286-293. DOI:10.1016/j.biortech.2018.03.065
|
| [13] |
YANG M, LU DW, QIN BD, LIU QL, ZHAO YM, LIU HL, MA J. Highly efficient nitrogen removal of a coldness-resistant and low nutrient needed bacterium, Janthinobacterium sp. M-11[J]. Bioresource Technology, 2018, 256: 366-373. DOI:10.1016/j.biortech.2018.02.049
|
| [14] |
SU JF, GUO L, YANG SF, ZHANG K, HUANG TL, WEN G. Heterotrophic nitrification and aerobic denitrification at low nutrient conditions by a newly isolated bacterium, Acinetobacter sp. SYF26[J]. Microbiology, 2015, 161(4): 829-837. DOI:10.1099/mic.0.000047
|
| [15] |
HUANG T, HE X, ZHANG H, ZHOU S, BAI S. Nitrogen removal characteristics of the heterotrophic nitrification-aerobic denitrification bacterium Acinetobacter sp. Sxf14[J]. Chinese Journal of Applied and Environmental Biology, 2015, 21: 201-207.
|
| [16] |
YU DY, GAO B, YANG MY, LIU WC, QIAO N. The treatment of heterotrophic nitrificationaerobic denitrifier bacteria loaded with bacterial cellulose membrane to nitrogenous wastewater[J]. Fresenius Environmental Bulletin, 2011, 20(5): 1208-1215.
|
| [17] |
陈杰, 汪霞, 赵彬, 安强, 黄源生. A. faecalis strain NR的包埋固定及其氨氮降解性能[J]. 环境工程学报, 2016, 10(6): 3303-3311. CHEN J, WANG X, ZHAO B, AN Q, HUANG YS. Embedding immobilization of A. faecalis strain NR and its capability of ammonium removal[J]. Chinese Journal of Environmental Engineering, 2016, 10(6): 3303-3311 (in Chinese). |
| [18] |
乔楠, 陈瑞佳, 于大禹. 硅藻土负载异养硝化-好氧反硝化菌的脱氮性能[J]. 化工进展, 2015, 34(5): 1459-1465. QIAO N, CHEN RJ, YU DY. Study on the denitrification characteristics of heterotrophic nitrification-aerobic denitrifier loaded on diatomite[J]. Chemical Industry and Engineering Progress, 2015, 34(5): 1459-1465 (in Chinese). |
| [19] |
张晨, 张培玉, 孙梦, 吴玉光. 菌株qy37吸附包埋固定化的脱氮效果研究[J]. 环境科学与技术, 2012, 35(1): 32-37. ZHANG C, ZHANG PY, SUN M, WU YG. Denitrification effect of heterotrophic nitrifying and aerobic denitrifying bacteria strain qy37 immobilized in absorbent and entrapping materials[J]. Environmental Science & Technology, 2012, 35(1): 32-37 (in Chinese). DOI:10.3969/j.issn.1003-6504.2012.01.007 |
| [20] |
赵立军, 马放, 山丹, 高敏. 曲霉菌丝球Y3对细菌的固定化效能[J]. 江苏大学学报(自然科学版), 2007, 28(5): 446-449. ZHAO LJ, MA F, SHAN D, GAO M. Immobilized effect of bacteria by mycelial pellet of Penicillum Y3[J]. Journal of Jiangsu University (Natural Science Edition), 2007, 28(5): 446-449 (in Chinese). DOI:10.3969/j.issn.1671-7775.2007.05.020 |
| [21] |
BOUCHEZ T, PATUREAU D, DELGENÈS JP, MOLETTA R. Successful bacterial incorporation into activated sludge flocs using alginate[J]. Bioresource Technology, 2009, 100(2): 1031-1032. DOI:10.1016/j.biortech.2008.07.028
|
| [22] |
WANG JL, CHU LB. Biological nitrate removal from water and wastewater by solid-phase denitrification process[J]. Biotechnology Advances, 2016, 34(6): 1103-1112. DOI:10.1016/j.biotechadv.2016.07.001
|
| [23] |
BOLEY A, MÜLLER WR, HAIDER G. Biodegradable polymers as solid substrate and biofilm carrier for denitrification in recirculated aquaculture systems[J]. Aquacultural Engineering, 2000, 22(1/2): 75-85.
|
| [24] |
HONDA Y, OSAWA Z. Microbial denitrification of wastewater using biodegradable polycaprolactone[J]. Polymer Degradation and Stability, 2002, 76(2): 321-327. DOI:10.1016/S0141-3910(02)00028-9
|
| [25] |
PATHAK VM, NAVNEET. Review on the current status of polymer degradation: a microbial approach[J]. Bioresources and Bioprocessing, 2017, 4(1): 1-31. DOI:10.1186/s40643-016-0134-4
|
| [26] |
汪鲁, 刘军, 李永富, 刘质浩, 崔鸿武. 基于固态碳源的同步硝化反硝化反应器对海水养殖废水中氮的去除性能[J]. 环境工程学报, 2022, 16(6): 2001-2009. WANG L, LIU J, LI YF, LIU ZH, CUI HW. Performance of simultaneous nitrification and denitrification reactor based on solid-phase carbon source on nitrogen removal from mariculture wastewater[J]. Chinese Journal of Environmental Engineering, 2022, 16(6): 2001-2009 (in Chinese). |
| [27] |
沈雯婷, 邱田娟, 何亦, 张树松. 一株好氧反硝化菌脱氮性能研究[J]. 广东化工, 2022, 49(14): 41-43. SHEN WT, QIU TJ, HE Y, ZHANG SS. Investigation into the denitrification efficiency of an aerobic denitrifying bacterium in a bioreactor system[J]. Guangdong Chemical Industry, 2022, 49(14): 41-43 (in Chinese). |
| [28] |
陈俊, 刘锋, 冯震, 耿雅雯, 李昂. PHBV异养/硫自养协同反硝化滤池强化硝酸盐去除[J]. 水处理技术, 2022, 48(5): 136-140. CHEN J, LIU F, FENG Z, GENG YW, LI A. Enhanced nitrate removal by PHBV heterotrophic/sulfur autotrophic cooperative denitrification filter[J]. Technology of Water Treatment, 2022, 48(5): 136-140 (in Chinese). DOI:10.16796/j.cnki.1000-3770.2022.05.028 |
| [29] |
ZHANG SS, SUN XB, FAN YT, QIU TL, GAO M, WANG XM. Heterotrophic nitrification and aerobic denitrification by Diaphorobacter polyhydroxybutyrativorans SL-205 using poly (3-hydroxybutyrate-co-3-hydroxyvalerate) as the sole carbon source[J]. Bioresource Technology, 2017, 241: 500-507. DOI:10.1016/j.biortech.2017.05.185
|
| [30] |
ZHAO TT, CHEN PP, ZHANG LJ, ZHANG L, GAO YH. Heterotrophic nitrification and aerobic denitrification by a novel Acinetobacter sp. TAC-1 at low temperature and high ammonia nitrogen[J]. Bioresource Technology, 2021, 339: 125620. DOI:10.1016/j.biortech.2021.125620
|
| [31] |
常慧敏, 杨青惠, 齐翔. TOC-L总有机碳分析仪测定总有机碳的实验方法[J]. 科技创新与生产力, 2017(11): 118-120. CHANG HM, YANG QH, QI X. Experimental method of total organic carbon determination based on TOC-L analyzer[J]. Taiyuan Science and Technology, 2017(11): 118-120 (in Chinese). DOI:10.3969/j.issn.1674-9146.2017.11.118 |
| [32] |
WANG LQ, QU FT, ZHU ZC, ZHAO Y, CHEN XM, SHI MZ, WEI ZM. The important role of tricarboxylic acid cycle metabolism pathways and core bacterial communities in carbon sequestration during chicken manure composting[J]. Waste Management, 2022, 150: 20-29. DOI:10.1016/j.wasman.2022.06.034
|
| [33] |
刘钊远. 增强地衣芽胞杆菌TCA循环代谢水平高产杆菌肽[D]. 武汉: 湖北大学硕士学位论文, 2018. LIU ZY. Enhance TCA cycle metabolism level of Bacillus licheniformis and high production of bacitracin[D]. Wuhan: Master's Thesis of Hubei University, 2018 (in Chinese). |
| [34] |
HENRISSAT B. A classification of glycosyl hydrolases based on amino acid sequence similarities[J]. The Biochemical Journal, 1991, 280(Pt 2): 309-316.
|
| [35] |
KOROPATKIN NM, CAMERON EA, MARTENS EC. How glycan metabolism shapes the human gut microbiota[J]. Nature Reviews Microbiology, 2012, 10(5): 323-335. DOI:10.1038/nrmicro2746
|
| [36] |
ZHU F, ZHANG H, WU H. Glycosyltransferase- mediated sweet modification in oral streptococci[J]. Journal of Dental Research, 2015, 94(5): 659-665. DOI:10.1177/0022034515574865
|
| [37] |
BIELY P. Microbial carbohydrate esterases deacetylating plant polysaccharides[J]. Biotechnology Advances, 2012, 30(6): 1575-1588. DOI:10.1016/j.biotechadv.2012.04.010
|
| [38] |
LOMBARD V, BERNARD T, RANCUREL C, BRUMER H, COUTINHO PM, HENRISSAT B. A hierarchical classification of polysaccharide lyases for glycogenomics[J]. The Biochemical Journal, 2010, 432(3): 437-444. DOI:10.1042/BJ20101185
|
| [39] |
CANTAREL BL, COUTINHO PM, RANCUREL C, BERNARD T, LOMBARD V, HENRISSAT B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics[J]. Nucleic Acids Research, 2009, 37(suppl_1): D233-D238.
|
| [40] |
JONES DR, THOMAS D, ALGER N, GHAVIDEL A, DOUGLAS INGLIS G, ABBOTT DW. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets[J]. Biotechnology for Biofuels, 2018, 11: 27. DOI:10.1186/s13068-018-1027-x
|
| [41] |
GHARECHAHI J, SALEKDEH GH. A metagenomic analysis of the camel rumen's microbiome identifies the major microbes responsible for lignocellulose degradation and fermentation[J]. Biotechnology for Biofuels, 2018, 11(1): 1-19. DOI:10.1186/s13068-017-1003-x
|
| [42] |
黄右琴. 湖羊瘤胃细菌基因组集构建及其碳水化合物活性酶挖掘[D]. 兰州: 兰州大学硕士学位论文, 2021. HUANG YQ. Construction of rumen bacterial genome and mining of carbohydrate active enzymes in Hu-sheep[D]. Lanzhou: Master's Thesis of Lanzhou University, 2021 (in Chinese). |
| [43] |
WANG HS, CHEN N, FENG CP, DENG Y. Insights into heterotrophic denitrification diversity in wastewater treatment systems: progress and future prospects based on different carbon sources[J]. Science of the Total Environment, 2021, 780: 146521. DOI:10.1016/j.scitotenv.2021.146521
|
| [44] |
GOLDBRAIKH D, NEUFELD D, EID-MUTLAK Y, LASRY I, GILDA JE, PARNIS A, COHEN S. USP1 deubiquitinates Akt to inhibit PI3K-Akt-FoxO signaling in muscle during prolonged starvation[J]. EMBO Reports, 2020, 21(4): e48791. DOI:10.15252/embr.201948791
|
| [45] |
YU L, WEI J, LIU PD. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer[J]. Seminars in Cancer Biology, 2022, 85: 69-94. DOI:10.1016/j.semcancer.2021.06.019
|
| [46] |
马云, 李婷婷, 冯日月, 郭桂杰, 潘启东, 李建宁, 陈吉龙. 急性白血病细胞中JAK/STAT5和PI3K/AKT信号通路对eIF4B的协同调控作用[J]. 生物工程学报, 2020, 36(11): 2413-2423. MA Y, LI TT, FENG RY, GUO GJ, PAN QD, LI JN, CHEN JL. Synergistic role of JAK/STAT5 and PI3K/AKT signaling pathways in regulating eIF4B in acute leukemia[J]. Chinese Journal of Biotechnology, 2020, 36(11): 2413-2423 (in Chinese). |
| [47] |
WANG C, CHEN YW, ZHOU HZ, LI XD, TAN ZL. Adaptation mechanisms of Rhodococcus sp. CNS16 under different temperature gradients: physiological and transcriptome[J]. Chemosphere, 2020, 238: 124571. DOI:10.1016/j.chemosphere.2019.124571
|
| [48] |
WU MX, ZOU Y, YU YH, CHEN BX, ZHENG QW, YE ZW, WEI T, YE SQ, GUO LQ, LIN JF. Comparative transcriptome and proteome provide new insights into the regulatory mechanisms of the postharvest deterioration of Pleurotus tuoliensis fruitbodies during storage[J]. Food Research International, 2021, 147: 110540. DOI:10.1016/j.foodres.2021.110540
|
| [49] |
FÜRCH T, HOLLMANN R, WITTMANN C, WANG W, DECKWER WD. Comparative study on central metabolic fluxes of Bacillus megaterium strains in continuous culture using 13C labelled substrates[J]. Bioprocess and Biosystems Engineering, 2007, 30(1): 47-59.
|
| [50] |
KWONG WK, ZHENG H, MORAN NA. Convergent evolution of a modified, acetate-driven TCA cycle in bacteria[J]. Nature Microbiology, 2017, 2: 17067. DOI:10.1038/nmicrobiol.2017.67
|
| [51] |
TANG KH, TANG YJ, BLANKENSHIP RE. Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications[J]. Frontiers in Microbiology, 2011, 2: 165.
|
| [52] |
ZHOU JW, LIU LM, SHI ZP, DU GC, CHEN J. ATP in current biotechnology: regulation, applications and perspectives[J]. Biotechnology Advances, 2009, 27(1): 94-101.
|
| [53] |
ATSUSHI Y, MASARU H, NAOHISA T, CHIHO H, YUJI T, YOSHIHIRO F, FUSAO T. Enhancement of glucose metabolism in a pyruvic acid-hyperproducing Escherichia coli mutant defective in F1-ATPase activity[J]. Journal of Fermentation and Bioengineering, 1997, 83(2): 132-138.
|
| [54] |
AOKI R, WADA M, TAKESUE N, TANAKA K, YOKOTA A. Enhanced glutamic acid production by a H+-ATPase-defective mutant of Corynebacterium glutamicum[J]. Bioscience, Biotechnology, and Biochemistry, 2005, 69(8): 1466-1472.
|
| [55] |
LIU LM, LI Y, LI HZ, CHEN J. Significant increase of glycolytic flux in Torulopsis glabrata by inhibition of oxidative phosphorylation[J]. FEMS Yeast Research, 2006, 6(8): 1117-1129.
|
| [56] |
AILEN M, SÁNCHEZ, GEORGE N. B, KA-YIU S. Effect of different levels of NADH availability on metabolic fluxes of Escherichia coli chemostat cultures in defined medium[J]. Journal of Biotechnology, 2005, 117(4): 395-405.
|
| [57] |
LIN H, CASTRO NM, BENNETT GN, SAN KY. Acetyl-CoA synthetase overexpression in Escherichia coli demonstrates more efficient acetate assimilation and lower acetate accumulation: a potential tool in metabolic engineering[J]. Applied Microbiology and Biotechnology, 2006, 71(6): 870-874.
|
 2023, Vol. 39
2023, Vol. 39




