
中国科学院微生物研究所、中国微生物学会主办
文章信息
- 杨文华, 顾秋亚, 余晓斌
- YANG Wenhua, GU Qiuya, YU Xiaobin
- 原人参二醇型皂苷水解酶及制备人参皂苷Compound K的研究进展
- Protopanaxadiol-type ginsenoside hydrolases and their application in the preparation of ginsenoside Compound K: a review
- 生物工程学报, 2023, 39(3): 978-992
- Chinese Journal of Biotechnology, 2023, 39(3): 978-992
- 10.13345/j.cjb.220492
-
文章历史
- Received: June 23, 2022
- Accepted: August 31, 2022
- Published: September 2, 2022
2. 江南大学生物工程学院, 江苏 无锡 214000
2. School of Biotechnology, Jiangnan University, Wuxi 214000, Jiangsu, China
人参是五加科的一种多年生草本植物,长久以来在全球被用于保持身体活力和延年益寿,素有“中药之王”的美称。大量文献表明,人参中的主要活性成分是人参皂苷,其在抗癌、抗炎和抗氧化等方面显示出巨大的应用价值[1-5]。截至目前,已有200多种人参皂苷及其衍生物被报道[3]。其中,达玛烷型人参皂苷由相似的达玛烷(四环三萜)骨架和糖基部分组成。由于苷元骨架上糖基的类型、附着位置以及数量的差异,达玛烷型人参皂苷又可分为原人参二醇型皂苷(Rb1、Rb2、Rb3、Rc、Rd、F2、CK等)和原人参三醇型皂苷(Re、Rf等)[6]。此外,根据去糖基化的程度,人参皂苷又可分为主要人参皂苷和稀有人参皂苷。稀有人参皂苷是通过主要人参皂苷去糖基化制备,在天然人参植物中不存在或者含量非常低[7]。其中,稀有人参皂苷(例如CK)因其具有比主要人参皂苷更高的生物利用度备受学者们的关注。
稀有人参皂苷CK,于1972年首次被发现,是原人参二醇型皂苷去糖基化的主要产物[8]。半个世纪以来,CK对癌症、肿瘤、炎症、糖尿病等多种疾病的(辅助)治疗能力已在许多研究中得到证实[9]。CK具有高度的安全性和多样化的生物学功能,可能是许多疾病的潜在治疗剂。目前,一种含有稀有人参皂苷CK的药片作为类风湿关节炎的候选药物,已在我国完成全部的0期和Ⅰ期临床试验[10]。随着CK市场需求量不断增大,如何大批量地生产这种在人参植株中不存在或者含量非常低的代谢物成为国内外学者们的研究方向。
以往的许多研究专注于使用热处理法、酸碱处理法、微生物转化法或酶促转化法将原人参二醇型主要人参皂苷转化为稀有人参皂苷[11-13]。其中,加热或酸碱处理法对于糖苷键的水解是随机的,导致产物的结构和类型难以预期。而且,尚无文献报道使用酸水解法制备CK。这可能是因为位于C-20的糖基在物理化学条件下可被非特异性裂解,而位于C-3的糖基则不能[9]。此外,物理化学法制备过程中产生的过高的能耗和废弃化合物对环境具有严重的污染性。微生物法为制备CK提供了一种环境友好型的方案。但是,菌株有限的生命力和较长的生长周期增加了工业生产的成本。相比较于这些方法,具有特异选择性的酶促催化法在绿色、稳定生产CK时具有显著优势[14]。为了满足人参皂苷CK不断增加的需求量和工业生产的低成本要求,对于更高效、更稳定以及底物适用范围更广泛的原人参二醇型皂苷水解酶的挖掘和研究仍然是迫切的。因此,本文主要就近年来原人参二醇型皂苷水解酶的发展状况及其在制备CK中的应用展开详述。
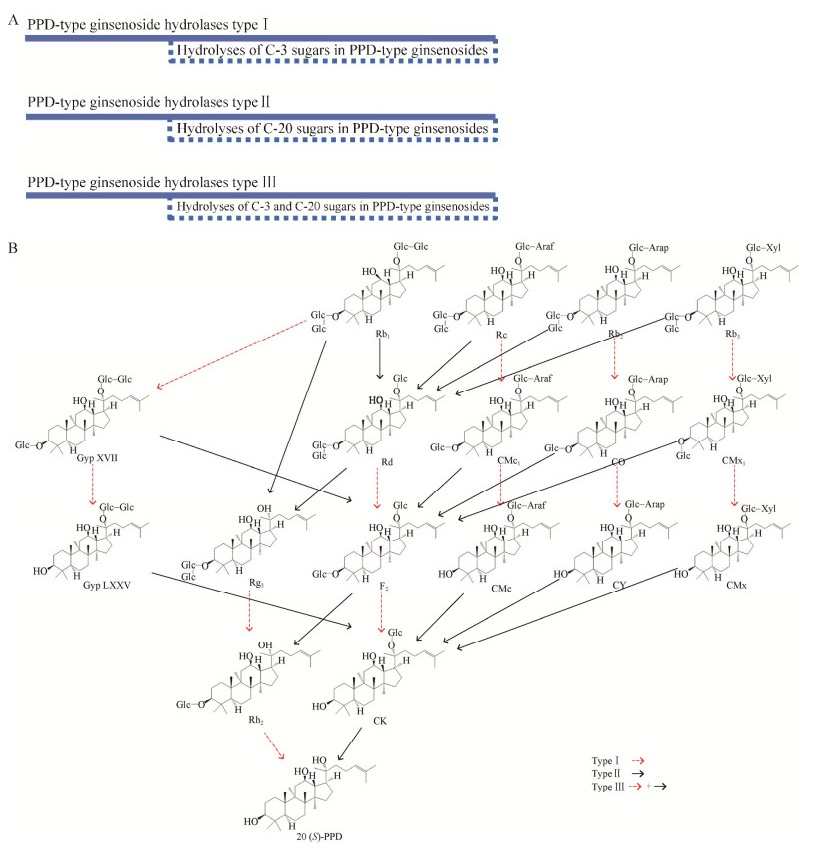
1 原人参二醇型皂苷水解酶原人参二醇型皂苷水解酶,是一种可以水解原人参二醇型皂苷的糖苷水解酶。以往的研究表明,原人参二醇型皂苷中涉及的糖基包含β-d-吡喃葡萄糖基、β-d-吡喃木糖基、α-l-阿拉伯呋喃糖基和α-l-阿拉伯呋喃糖基,通过C-3和/或C-20与原人参二醇型苷元进行连接(化学结构如表 1所示)[6]。截至目前,对原人参二醇型皂苷水解酶分类的研究较少[15-16]。以往对原人参二醇型皂苷水解酶的分类是依据水解酶作用的糖基与苷元骨架连接还是与糖基连接,把原人参二醇型皂苷水解酶分为6–8种。为了便于理解与归类,我们根据水解酶作用的糖基连接的碳原子的不同,将原人参二醇型皂苷水解酶分为3种类型(图 1A)。其中,仅能水解C-3上糖基的水解酶被定义为Ⅰ型原人参二醇型皂苷水解酶。仅能水解C-20上糖基的水解酶被定义为Ⅱ型原人参二醇型皂苷水解酶。能同时水解C-3和C-20上糖基的水解酶被定义为Ⅲ型原人参二醇型皂苷水解酶。图 1B显示了原人参二醇型皂苷之间的转化关系。表 2列出了近几年挖掘的原人参二醇型皂苷水解酶,这些水解酶来源于不同的菌株,最适催化条件也有较大差别。
| Chemical structure | PPD-type ginsenosides | R1 | R2 |
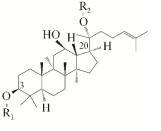 |
Rb1 | Glc(2→1)Glc | Glc(6→1)Glc |
| Rd | Glc(2→1)Glc | Glc | |
| F2 | Glc | Glc | |
| Rb2 | Glc(2→1)Glc | Glc(6→1)Arap | |
| CO | Glc | Glc(6→1)Arap | |
| CY | H | Glc(6→1)Arap | |
| Rb3 | Glc(2→1)Glc | Glc(6→1)Xyl | |
| CMx1 | Glc | Glc(6→1)Xyl | |
| CMx | H | Glc(6→1)Xyl | |
| Rc | Glc(2→1)Glc | Glc(6→1)Araf | |
| CMc1 | Glc | Glc(6→1)Araf | |
| CMc | H | Glc(6→1)Araf | |
| Gyp XVII | Glc | Glc(6→1)Glc | |
| Gyp LXXV | H | Glc(6→1)Glc | |
| CK | H | Glc | |
| Rg3 | Glc(2→1)Glc | H | |
| Rh2 | Glc | H | |
| 20 (S)-PPD | H | H | |
| Glc: β-d-glucopyranosyl; Arap: α-l-arabinopyranosyl; Araf: α-l-arabinofuranosyl; Xyl: β-d-xylopyranyl. | |||
|
| 图 1 原人参二醇型皂苷水解酶的分类(A)及原人参二醇型皂苷的转化途径(B) Fig. 1 Classification of PPD-type ginsenoside hydrolases (A) and transformation pathway of PPD-type ginsenosides (B). |
| |
| Type | Protein | GH family | Strain | pH | Tm (℃) | Time (h) | Enzyme concentration | Ginsenoside | Yield | Transformationpathway | References |
| Ⅰ | β-glucosidase | 3 | Bifidobacterium breve ATCC 15700 | 5.0 | 35 | 12.0 | 6 U/mL | 5 mmol/L Rd | 4.35 mmol/L CK, 100% | Rd→F2→CK | [17] |
| β-glucosidase | 3 | Shingopyxis alaskensis | 5.5 | 40 | 1.0 | 0.5 mg/mL | 8 mg/mL Rb1 | 6.8 mg/mL Gyp XVII | Rb1→Gyp XVII | [18] | |
| β-glucosidase | 1 | Cellulosimicrobium cellulans sp. 21 | 5.5 | 35 | 2.0 | 1 mg/mL | 1 g PPDGM | 292 mg Gyp XVII, 134 mg CO, 184 mg CMc1, and 62 mg F2 | Rb1→Gyp XVII, Rb2→CO, Rc→CMc1, Rd→F2 | [19] | |
| Ⅱ | β-glucosidase | 3 | Microbacterium esteraromaticum | 7.0 | 37 | 1.0 | 0.75 U/mL | 10 mg/mL Rb1 | 6.9 mg/mL Rg3, 97.8% | Rb1→Rd→Rg3 | [20] |
| β-glycosidase | 2 | Microbacterium esteraromaticum GS514 | 7.0 | 40 | 2.0 | 0.1 mg/mL | 1 mg/mL Rb2 | 0.47 mg/mL Rg3 | Rb2→Rd→Rg3 | [21] | |
| β-glucosidase | – | Flavobacterium johnsoniae | 7.0 | 30 | 24.0 | 0.001 U/mL | 3.4 mmol/L Rb1, and 2.4 mmol/L Rd | 61% Rg3 | Rb1→Rd→Rg3 | [22] | |
| β-glycosidase | 1 | Thermotoga petrophlia | 5.0 | 90 | 3.0 | 1.2 U/mL | 10 mg/mL ginsenoside extract | 3.93 mg/mL Rg3, 98.19% | Rb1 or Rb2→Rd→Rg3 | [23] | |
| α-L-arabinofuranosidase | 51 | Thermotoga thermarum DSM5069 | 5.0 | 85 | 1.0 | 12 U/mL | 25 mg/mL Rc | 21·8 mg/mL Rd, 99.4% | Rc→Rd | [24] | |
| Ⅲ | β-glucosidase | 3 | Microbacterium esteraromaticum | 7.0 | 40 | 1.0 | 0.1 mg/mL | 1 mg/mL Rb1 | 0.46 mg/mL CK, 77% | Rb1→Rd→CK | [25] |
| β-glycosidase | – | Sulfolobus solfataricus | 5.5 | 85 | 12.0 | 40 U/mL | 1.9 mg/mL Rb1, 0.52 mg/mL Rb2, 0.92 mg/mL Rc, and 0.23 mg/mL Rd | 1.63 mg/mL CK | Rb1 or Rb2→Rd→F2→CK, and Rc →CMc→CK | [26] | |
| β-glycosidase | – | Sulfolobus solfataricus | 5.5 | 95 | 1.5 | 0.01 mg/mL | 2 mg/mL Rb1 | 1.12 mg/mL CK, 100% | Rb1→Rd→F2→CK | [27] | |
| β-glucosidase | 3 | Actinosynnema mirum KACC 20028T | 8.0 | 30 | – | 1 mg/mL | 0.1% (W/V) Rb1, Rb2, Rc, Rd, Rg3 | – | Rb1→Gyp XVII→Gyp LXXV, Rh2→PPD, Rb2→CO→CY, Rc→CMc1→CMc, Rd→F2→Rh2→PPD, Rg3→Rh2→PPD | [28] | |
| Lactase | – | Aspergillus oryzae | 4.5 | 37 | 72.0 | 2 g | 0.4 g of PPD-type saponin | – | Rb1, Rb2, Rb3→Rd→Rg3, Rb1, Rb2, Rb3, Rc→Rd→F2→CK | [29] |
如图 1B所示,用虚线箭头指示的转化途径需要用Ⅰ型原人参二醇型皂苷水解酶进行催化。
Zhang等报道了一种来自短双歧杆菌(Bifidobacterium breve) ATCC 15700的GH3家族的β-葡萄糖苷酶,该Ⅰ型水解酶可在12 h内将5 mmol/L Rd全部转化为CK,能够逐步水解连在Rd的C-3上的内外两个β-d-吡喃葡萄糖基[17]。Shin等发现一种来自阿拉斯加鞘氨醇盒菌(Sphingopyxis alaskensis)的GH3家族的β-葡萄糖苷酶,该Ⅰ型水解酶可在1 h内将8 mg/mL Rb1转化为6.8 mg/mL Gypenoside XVII (Gyp XVII),可逐步水解连在Rb1的C-3上的内外两个β-d-吡喃葡萄糖基[18]。Yuan等发现一种来自纤维纤维微菌(Cellulosimicrobium cellulans) 21的GH1家族的β-葡萄糖苷酶,该Ⅰ型水解酶可在2 h内将1 g原人参二醇型皂苷转化为292 mg Gyp XVII、134 mg CO、184 mg CMc1和62 mg F2,可水解连在C-3上的内外两个β-d-吡喃葡萄糖基[19]。
综上所述,Ⅰ型原人参二醇型皂苷水解酶在酶量和时间充足的情况下,可将C-3上连接的内外β-d-吡喃葡萄糖基逐步水解。
1.2 Ⅱ型原人参二醇型皂苷水解酶如图 1B所示,用实线箭头指示的转化途径需要用Ⅱ型原人参二醇型皂苷水解酶进行催化。
Quan等报道一种来自酯香微杆菌(Microbacterium esteraromaticum)的GH3家族的β-葡萄糖苷酶,该Ⅱ型水解酶在1 h内能将10 mg/mL Rb1转化为6.9 mg/mL Rg3,可逐步水解C-20上的内外两个β-d-吡喃葡萄糖基[20]。Quan等发现一种来自M. esteraromaticum GS514的GH2家族的β-糖苷酶,该Ⅱ型水解酶可在2 h内将1 mg/mL Rb2转化为0.47 mg/mL Rg3,水解连在C-20上的α-l-阿拉伯呋喃糖基和β-d-吡喃葡萄糖基[21]。Li等发现一种来自约氏黄杆菌(Flavobacterium johnsoniae)的β-葡萄糖苷酶,该Ⅱ型水解酶在24 h内能将Rb1转化为Rg3 (摩尔转化率为61%),可逐步水解C-20上的内外两个β-d-吡喃葡萄糖基[22]。Zhang等报道一种来自超嗜热菌(Thermotoga petrophlia)的GH1家族的β-糖苷酶,该Ⅱ型水解酶可在3 h内将10 mg/mL人参提取物转化为3.93 mg/mL Rg3,可逐步水解C-20上的内外两个β-d-吡喃葡萄糖基[23]。Xie等发现一种来自嗜热菌(Thermotoga thermarum) DSM5069的GH51家族的α-l-阿拉伯呋喃糖苷水解酶,该Ⅱ型水解酶可在1 h将25 mg/mL Rc转化为21.8 mg/mL Rd,水解C-20上的α-l-阿拉伯呋喃糖基[24]。
综上所述,Ⅱ型原人参二醇型皂苷水解酶可作用于C-20上连接的β-d-吡喃葡萄糖基、α-l-阿拉伯呋喃糖基、α-l-阿拉伯吡喃糖基和β-d-吡喃木糖基。此外,从类似M. esteraromaticum等具有转化效力的微生物中挖掘出多种Ⅱ型原人参二醇型皂苷水解酶。
1.3 Ⅲ型原人参二醇型皂苷水解酶如图 1B所示,包含两种类型箭头的转化途径需要用Ⅲ型原人参二醇型皂苷水解酶进行催化。
Quan等报道了一种来自M. esteraromaticum的GH3家族的β-葡萄糖苷酶,该Ⅲ型水解酶在1 h内能将1 mg/mL Rb1转化为0.46 mg/mL CK,可水解Rb1的C-3和C-20上的糖基生成CK[25]。Noh等发现一种来自硫磺矿硫化叶菌(Sulfolobus solfataricus)的β-糖苷酶,该Ⅲ型水解酶在12 h内将1.9 mg/mL Rb1、0.52 mg/mL Rb2、0.92 mg/mL Rc和0.23 mg/mL Rd转化为1.63 mg/mL CK,可同时水解Rb1、Rb2、Rc和Rd的C-3和C-20上的糖基生成CK[26]。Shin等报道了一种来自S. solfataricus的β-糖苷酶,该Ⅲ型水解酶在1.5 h内能将2 mg/mL Rb1完全转化为1.12 mg/mL CK,可水解Rb1的C-3和C-20上的糖基生成CK[27]。Cui等发现一种来自奇迹束丝放线菌(Actinosynnema mirum) KACC 20028T的β-葡萄糖苷酶,该Ⅲ型水解酶可将天然皂苷Rb1、Rb2、Rc和Rd转化为稀有人参皂苷Rg3、CK和20 (S)-PPD,水解多种主要人参皂苷的C-3和C-20上的糖基生成稀有人参皂苷[28]。Ko等发现一种来自米曲霉(Aspergillus oryzae)的乳糖酶,该Ⅲ型水解酶可将天然皂苷Rb1、Rb2、Rc、Rb3和Rd转化为稀有人参皂苷Rg3、CK和20 (S)-PPD,水解多种皂苷的C-3和C-20上的糖基生成稀有人参皂苷[29]。
综上所述,Ⅲ型原人参二醇型皂苷水解酶可作用于C-3和C-20上连接的β-d-吡喃葡萄糖基、α-l-阿拉伯呋喃糖基、α-l-阿拉伯吡喃糖基和β-d-吡喃木糖基。此外,从类似M. esteraromaticum和S. solfataricus等具有转化效力的微生物中挖掘出多种Ⅲ型原人参二醇型皂苷水解酶。
2 原人参二醇型皂苷水解酶在CK生产上的应用一个多世纪以来,酶在工业中的使用引起了人们的关注,工业酶的全球市场不断增长,预计在2022年可达到85亿美元的市场[16]。这种绿色、高效且特异性强的生物催化剂为生产增值化合物提供了一个综合性的强大方法库。基于对原人参二醇型皂苷水解酶的分类,近几年水解酶在CK生产上的案例也被总结归纳。
2.1 天然酶水解制备CK有氧条件下的微生物转化为人参皂苷的生产提供了一种高效且廉价的工艺。究其根本,具有人参皂苷转化效力的微生物具有特异性的酶催化剂,可以水解原人参二醇型皂苷生产CK。
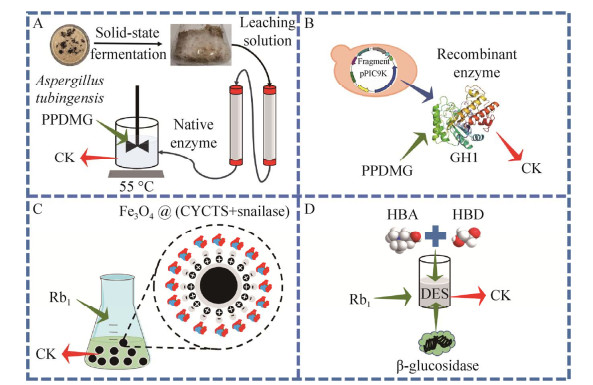
拟青霉(Paecilomyces bainier) 229的胞外酶经分离纯化得到7种组分,每种组分都具有水解人参皂苷的能力,其中一种被定义为β-葡萄糖苷酶的组分可特异性地将Rb1水解为CK[30]。Chang等报道了一株黑曲霉(Aspergillus niger)的胞外酶可将Rb1水解为Rg3和CK[31]。来源于蜜环菌(Armillaria mellea)菌丝体的胞内酶与1 mg/mL的Rb1和Rb2反应96 h,皂苷被全部水解为CK[32-33]。Kim等从发酵大豆砖中分离出一种可生产CK的塔宾曲霉(Aspergillus tubingensis),其经分离纯化的胞外β-葡萄糖苷酶与原人参二醇型皂苷(包含Rb1、Rb2、Rb3和Rc)反应20 h后,可将13.4 mmol/L原人参二醇型皂苷完全转化为CK (图 2A),这是第一个将所有原人参二醇型皂苷完全转化为CK的食品安全(generally recognized as safe, GRAS) 酶[34]。除此之外,多项研究报道了微生物天然酶对人参皂苷的水解能力[35-39],微生物资源丰富,是尚待深入挖掘新酶的宝库。
|
| 图 2 天然酶(A)、重组酶(B)、固定化酶(C)、溶剂工程在CK生产中的应用(D) Fig. 2 Application of native enzymes (A), recombinant enzymes (B), immobilized enzymes (C), and solvent engineering (D) in the production of CK. |
| |
所筛选的微生物通过发酵的方式被大量制备,这些菌体被处理后得到大量的天然生物催化剂。生物催化剂通过纯化的方式,得到纯度较高的酶制剂。但是,天然酶制剂的纯化制备过程复杂,需要耗费大量的人力物力。随着基因工程技术的不断发展,可通过克隆表达的方式大量制备具有水解特异型的酶制剂,且表达产物具有纯度高、活性强的优点。
2.2 重组酶水解制备CK研究表明[40-45],大多数能制备CK的水解酶为Ⅲ型原人参二醇型皂苷水解酶,而且来自GH1和GH3家族的β-葡萄糖苷酶对原人参二醇型皂苷的水解作出了巨大贡献(表 1)。
来自黄杆菌(Flavobacterium chilense)的GH3家族的β-葡萄糖苷酶在大肠杆菌(Escherichia coli) BL21中成功表达[46],该酶可用于制备CK,且最适温度为37 ℃、最适pH 7.0。但是,该酶在温度超过45 ℃时,酶活性仅能维持甚至低于初始酶活性的20%。较高的酶促反应温度会促进水溶性较差底物(例如,大多数人参皂苷)在反应液中的溶解度,这种正向的促进作用会加快底物与酶制剂之间的传质速率从而降低生产的时间成本。相似地,在E. coli BL21中成功表达的β-葡萄糖苷酶可耐受高达40 g/L的人参皂苷Rd[17]。这种高浓度的底物耐受性有利于在工业生产中降低酶制剂的使用成本。此外,在谷氨酸棒状杆菌(Corynebacterium glutamicum) ATCC 13032中表达的重组C3a-MT619,仅具有在E. coli BL21中异源表达的MT619的61.9%的酶活性[7]。因此,不同的表达系统对于同种酶的表达效果并不相同。来源于尖海龙共附生真菌(Talaromyces amestolkiae)的β-葡萄糖苷酶在毕赤酵母(Pichia pastoris)中被高效表达,75 U/mL的生产水平是文献中报道的重组β-葡萄糖苷酶的较高水平[47]。由此得出,在筛选最优表达系统时,表达系统的生物安全水平、表达量以及对基因编码的偏好性等都是需要评估的条件。此外,挖掘报道的水解酶多用于水解单一底物,对于酶的底物适用范围应该被扩大研究。新的生物催化剂在绿色化学和可持续化学工程中占有重要地位,有望克服现有生物催化剂的不足[48]。那些被报道的具有转化人参皂苷效果的微生物,是发现新型生物催化剂的巨大宝库(图 2B)。未来,学者们对于新型生物催化剂的持续开发,是推动稀有人参皂苷产业不断前进的基础且关键的一步。
此外,人参皂苷结构多样,不同类型的人参皂苷水解酶可特异性水解人参皂苷中的不同的糖基部分[16]。不同类型的人参皂苷水解酶组合使用策略为实现CK的高效生产提供了参考[15]。基于BglSk与BglPm的组合酶催化策略可制得(277.61±4.27) mg的CK,高于单一酶促反应的产量[49]。还有研究表明,可以通过随机突变或者理性设计的方式改变蛋白质的氨基酸序列,从而改善蛋白的理化能力和催化性能[50]。通过对来源于S. solfataricus的β-糖苷酶进行理性设计,CK的产率由306 mg/(L·h)提升至843 mg/(L·h)[27]。Shin等对源于番茄溃疡病菌(Clavibacter michiganensis)的β-葡萄糖苷酶512位氨基酸残基进行了靶向诱变,拓宽了突变体的区域选择性,使变体酶具有了将Rb1转化为CK的能力(以前不能)[51]。这种基于酶的定向进化的大量变体库生成后,可筛选和选择表现出所需特征的最佳变体[52]。未来,可以通过蛋白质工程优化相关蛋白的催化性能和理化功能,为CK的制备降低酶制剂成本。
2.3 固定化酶水解制备CK异源表达的方式使短期内大量制备纯度高、特异性强的酶制剂成为可能。但是,在实际的工业生产中,如何稳定发挥酶的催化性能以及重复使用酶制剂仍是有待解决的问题[53]。因此,易于回收且能维持酶稳定性和活性的固定化酶受到研究者们的关注。这也为重复、高效使用酶制剂催化生产CK提供了参考。
使用GRAS菌株C. glutamicum ATCC 13032表达了一种来源于M. testaceum且融合纤维素结合模块的新型β-葡萄糖苷酶[7]。融合蛋白以高密度(最大984 mg/g纤维素)有效地固定在无定形纤维素上,使酶浓度提高了286倍。高浓度的固定化酶可在24 h内制备7.59 g/L CK。Shi等将GH3家族的β-葡萄糖苷酶(Tpebgl3)负载在Fe3O4-SiO2-NH2-Cellu-ZIF8上,这种金属有机骨架磁性颗粒赋予了Tpebgl3更优异的高温稳定性和耐酸性[54],而且在10个重复循环结束时,固定化酶还保留了74%的初始活性。Aslihan等研究制备了一种基于藻酸盐的固定化β-葡萄糖苷酶的酶微反应器,该系统在连续流动条件下运行,可持续将Rb1水解为CK[55]。Li等合成了50 nm的球形Fe3O4@CYCTS纳米粒子[56]。如图 2C所示,这种用羧化壳聚糖包封磁铁矿制备的纳米颗粒为蜗牛酶的固定提供了大的比表面积,固定化的蜗牛酶可用于连续生产CK,在9个连续循环后固定化酶仍保留了约56%的初始催化活性。Wu等将来自A. niger的β-葡萄糖苷酶通过化学亲和的方法固定在氨基二氧化硅中,固定化酶拥有比游离酶更宽的pH范围和更高的热稳定性,并且在重复使用制备CK的45个循环后仍可保持97%的初始活性[57]。
为了扩大酶制剂的工业化应用范围,越来越多的研究专注于探索新的固定基质和策略[58-62]。但是,常见的固定化策略多专注于单一蛋白的固定。未来,应该考虑多种蛋白的组合固定策略。
2.4 溶剂工程用于水解制备CK在生物催化的过程中,固定化酶可以起到改善酶性质的作用。但是,对于大多数合成反应来说,疏水性底物(例如,大多数人参皂苷)在反应溶剂中的溶解度也是影响反应进程的一大问题[63]。有机溶剂在一定程度上可以促进疏水性底物的溶解性,但是与水溶液相比,大多数酶制剂在有机溶剂中的催化效率较低。随着溶剂工程的不断发展,低共熔溶剂(deep eutectic solvent, DES)因其低毒性、生物降解性、可回收性、易燃性和价格低廉逐渐引起学者们的关注[64]。已有研究表明,低共熔溶剂可用于疏水性化合物的浸提以及生物催化反应[65-69]。
相对于需要大量的有机溶剂、较长的提取时间和较高能量的传统提取方法,Jeong等研究了一种包含甘油、l-脯氨酸和蔗糖的DES,可以从人参提取物中低毒、绿色提取大量人参皂苷[70]。Li等开发了一种基于氯化胆碱、1, 4-丁二醇和无机盐溶液(K2HPO4)的水相两相系统,可以将中药注射剂(康爱注射液)中超过90%的人参皂苷(包括人参皂苷Rg1、Re、Rf、Rg2、Rb1、Rc、Rb2和Rd)进行提取回收[71]。Han等建立的基于31.9% (质量分数) DES和24.5% (质量分数) K2HPO4的水相两相体系,可从顶部和底部相中回收75.79%的催化产物CK和61.14%的β-葡萄糖苷酶,被回收的β-葡萄糖苷酶可再次循环用于CK的制备[72]。这是首次将低共熔溶剂用于生物催化生产CK的研究。如图 2D所示,Ma等开发了一种基于氯化胆碱和乙二醇的DES,在30% (体积分数) DES存在的情况下,β-葡萄糖苷酶的半衰期增加了96%,F2的溶解度增加了120%,CK的产率增加了54%,在60 ℃、pH 4.5的条件下,8 mmol/L Rb1可转化为CK,转化率达到80.6%[73]。
DES通过季铵盐或金属盐与氢键供体的络合获得[74],已有多项研究证明其在生物催化领域的优势[75-79],为高效生产CK提供了可行的参考。此外,在实际的应用中,多组分络合形成的DES的生物毒性也应该被重视[80]。
3 总结与展望微生物来源的原人参二醇型皂苷水解酶,具有来源广泛、高催化效率和性质多样等特点,越来越多的原人参二醇型皂苷水解酶被挖掘并被用于生产稀有人参皂苷。相对于使用传统的物理化学手段制备CK,酶催化法不但生产工艺绿色环保,而且产量稳定高效。但是,原人参二醇型皂苷水解酶要应用于低成本、高产值的CK工业化生产时,还需要在以下4个方面不断改进。(1) 继续从自然界筛选具有耐高温、耐高浓度底物、底物适用范围更广泛的原人参二醇型皂苷水解酶。(2) 开发适用蛋白表达范围更广泛的表达系统,达到蛋白的最大表达产量,一定程度减少酶促转化制备CK的酶制剂成本。(3) 协同使用几种酶制剂或者制备方法,开发更优的制备策略,已达到CK产量的更大值。(4) 通过理性设计或代谢调控提高蛋白的表达量和活性,甚至赋予相关蛋白更优良的催化性能。
此外,随着生物技术的不断革新,可以利用合成生物学的手段将廉价的碳源(例如葡萄糖或甘油)在细胞体内加工为稀有人参皂苷[81-82]。而且,Wang等构建的工程酵母菌株在补料分批发酵中产生的CK滴度可达到5.74 g/L,这与5年生人参根的总皂苷含量(约3%)相当[83]。相较于传统的制备方法,这种更廉价、省力的生产方式是植物药用代谢物工业生产中的福音。未来,对于新兴技术的探索也在继续。
| [1] |
MAI TT, MOON J, SONG Y, VIET PQ, PHUC PV, LEE JM, YI TH, CHO M, CHO SK. Ginsenoside F2 induces apoptosis accompanied by protective autophagy in breast cancer stem cells. Cancer Letters, 2012, 321(2): 144-153. DOI:10.1016/j.canlet.2012.01.045
|
| [2] |
KIM JH. Pharmacological and medical applications of Panax ginseng and ginsenosides: a review for use in cardiovascular diseases. Journal of Ginseng Research, 2018, 42(3): 264-269. DOI:10.1016/j.jgr.2017.10.004
|
| [3] |
CHEN YY, LIU QP, AN P, JIA M, LUAN X, TANG JY, ZHANG H. Ginsenoside Rd: a promising natural neuroprotective agent. Phytomedicine, 2022, 95: 153883. DOI:10.1016/j.phymed.2021.153883
|
| [4] |
KIM E, KIM D, YOO S, HONG YH, HAN SY, JEONG S, JEONG D, KIM JH, CHO JY, PARK J. The skin protective effects of compound K, a metabolite of ginsenoside Rb1 from Panax ginseng. Journal of Ginseng Research, 2018, 42(2): 218-224. DOI:10.1016/j.jgr.2017.03.007
|
| [5] |
ZHANG JM, WANG YJ, JIANG YY, LIU TW, LUO YY, DIAO EJ, CAO YF, CHEN L, ZHANG L, GU Q, ZHOU JY, SUN FT, ZHANG WC, LIU JX, LI XQ, HU WC. Enhanced cytotoxic and apoptotic potential in hepatic carcinoma cells of chitosan nanoparticles loaded with ginsenoside compound K. Carbohydrate Polymers, 2018, 198: 537-545. DOI:10.1016/j.carbpol.2018.06.121
|
| [6] |
SHIN BK, KWON SW, PARK JH. Chemical diversity of ginseng saponins from Panax ginseng. Journal of Ginseng Research, 2015, 39(4): 287-298. DOI:10.1016/j.jgr.2014.12.005
|
| [7] |
CUI CH, JEON BM, FU YY, IM WT, KIM SC. High-density immobilization of a ginsenoside-transforming β-glucosidase for enhanced food-grade production of minor ginsenosides. Applied Microbiology and Biotechnology, 2019, 103(17): 7003-7015. DOI:10.1007/s00253-019-09951-4
|
| [8] |
LIU JL, WANG YC, YU Z, LV GF, HUANG XW, LIN H, MA C, LIN Z, QU P. Functional mechanism of ginsenoside compound K on tumor growth and metastasis. Integrative Cancer Therapies, 2022, 21: 15347354221101203.
|
| [9] |
YANG XD, YANG YY, OUYANG DS, YANG GP. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia, 2015, 100: 208-220. DOI:10.1016/j.fitote.2014.11.019
|
| [10] |
CHEN LL, ZHOU LP, HUANG J, WANG YQ, YANG GP, TAN ZR, WANG YC, ZHOU G, LIAO JW, OUYANG DS. Single- and multiple-dose trials to determine the pharmacokinetics, safety, tolerability, and sex effect of oral ginsenoside compound K in healthy Chinese volunteers. Frontiers in Pharmacology, 2018, 8: 965. DOI:10.3389/fphar.2017.00965
|
| [11] |
JANG GY, KIM MY, LEE YJ, LI MS, SHIN YS, LEE J, JEONG HS. Influence of organic acids and heat treatment on ginsenoside conversion. Journal of Ginseng Research, 2018, 42(4): 532-539. DOI:10.1016/j.jgr.2017.07.008
|
| [12] |
ZHANG R, TAN SQ, ZHANG BL, GUO ZY, TIAN LY, WENG P, LUO ZY. Two key amino acids variant of α-l-arabinofuranosidase from Bacillus subtilis str. 168 with altered activity for selective conversion ginsenoside Rc to Rd. Molecules (Basel, Switzerland), 2021, 26(6): 1733. DOI:10.3390/molecules26061733
|
| [13] |
LIU Z, XIA J, WANG CZ, ZHANG JQ, RUAN CC, SUN GZ, YUAN CS. Remarkable impact of acidic ginsenosides and organic acids on ginsenoside transformation from fresh ginseng to red ginseng. Journal of Agricultural and Food Chemistry, 2016, 64(26): 5389-5399. DOI:10.1021/acs.jafc.6b00963
|
| [14] |
LU CW, YIN YG. Pulsed electric field treatment combined with commercial enzymes converts major ginsenoside Rb1 to minor ginsenoside Rd. Innovative Food Science & Emerging Technologies, 2014, 22: 95-101.
|
| [15] |
SHIN KC, OH DK. Classification of glycosidases that hydrolyze the specific positions and types of sugar moieties in ginsenosides. Critical Reviews in Biotechnology, 2016, 36(6): 1036-1049. DOI:10.3109/07388551.2015.1083942
|
| [16] |
LI WN, FAN DD. Biocatalytic strategies for the production of ginsenosides using glycosidase: current state and perspectives. Applied Microbiology and Biotechnology, 2020, 104(9): 3807-3823. DOI:10.1007/s00253-020-10455-9
|
| [17] |
ZHANG R, HUANG XM, YAN HJ, LIU XY, ZHOU Q, LUO ZY, TAN XN, ZHANG BL. Highly selective production of compound K from ginsenoside Rd by hydrolyzing glucose at C-3 glycoside using β-glucosidase of Bifidobacterium breve ATCC 15700. Journal of Microbiology and Biotechnology, 2019, 29(3): 410-418. DOI:10.4014/jmb.1808.08059
|
| [18] |
SHIN KC, OH DK. Characterization of a novel recombinant β-glucosidase from Sphingopyxis alaskensis that specifically hydrolyzes the outer glucose at the C-3 position in protopanaxadiol-type ginsenosides. Journal of Biotechnology, 2014, 172: 30-37. DOI:10.1016/j.jbiotec.2013.11.026
|
| [19] |
YUAN Y, HU YB, HU CX, LENG JY, CHEN HL, ZHAO XS, GAO J, ZHOU YF. Overexpression and characterization of a glycoside hydrolase family 1 enzyme from Cellulosimicrobium cellulans sp. 21 and its application for minor ginsenosides production. Journal of Molecular Catalysis B: Enzymatic, 2015, 120: 60-67. DOI:10.1016/j.molcatb.2015.06.015
|
| [20] |
QUAN LH, MIN JW, YANG DU, KIM YJ, YANG DC. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant β-glucosidase from Microbacterium esteraromaticum. Applied Microbiology and Biotechnology, 2012, 94(2): 377-384. DOI:10.1007/s00253-011-3861-7
|
| [21] |
QUAN LH, WANG C, JIN Y, WANG TR, KIM YJ, YANG DC. Isolation and characterization of novel ginsenoside-hydrolyzing glycosidase from Microbacterium esteraromaticum that transforms ginsenoside Rb2 to rare ginsenoside 20(S)-Rg3. Antonie Van Leeuwenhoek, 2013, 104(1): 129-137. DOI:10.1007/s10482-013-9933-1
|
| [22] |
LI L, LEE SJ, YUAN QP, IM WT, KIM SC, HAN NS. Production of bioactive ginsenoside Rg3 (S) and compound K using recombinant Lactococcus lactis. Journal of Ginseng Research, 2018, 42(4): 412-418. DOI:10.1016/j.jgr.2017.04.007
|
| [23] |
ZHANG S, LUO J, XIE J, WANG Z, XIAO W, ZHAO L. Cooperated biotransformation of ginsenoside extracts into ginsenoside 20 (S)-Rg3 by three thermostable glycosidases. Journal of Applied Microbiology, 2020, 128(3): 721-734. DOI:10.1111/jam.14513
|
| [24] |
XIE J, ZHAO D, ZHAO L, PEI J, XIAO W, DING G, WANG Z, XU J. Characterization of a novel arabinose-tolerant α-l-arabinofuranosidase with high ginsenoside Rc to ginsenoside Rd bioconversion productivity. Journal of Applied Microbiology, 2016, 120(3): 647-660. DOI:10.1111/jam.13040
|
| [25] |
QUAN LH, MIN JW, JIN Y, WANG C, KIM YJ, YANG DC. Enzymatic biotransformation of ginsenoside Rb1 to Compound K by recombinant β-glucosidase from Microbacterium esteraromaticum. J Agric Food Chem, 2012, 60(14): 3776-3781. DOI:10.1021/jf300186a
|
| [26] |
NOH KH, SON JW, KIM HJ, OH DK. Ginsenoside compound K production from ginseng root extract by a thermostable β-glycosidase from Sulfolobus solfataricus. Bioscience, Biotechnology, and Biochemistry, 2009, 73(2): 316-321. DOI:10.1271/bbb.80525
|
| [27] |
SHIN KC, CHOI HY, SEO MJ, OH DK. Improved conversion of ginsenoside Rb1 to compound K by semi-rational design of Sulfolobus solfataricus β-glycosidase. AMB Express, 2017, 7(1): 186. DOI:10.1186/s13568-017-0487-x
|
| [28] |
CUI CH, KIM SC, IM WT. Characterization of the ginsenoside-transforming recombinant β-glucosidase from Actinosynnema mirum and bioconversion of major ginsenosides into minor ginsenosides. Applied Microbiology and Biotechnology, 2013, 97(2): 649-659. DOI:10.1007/s00253-012-4324-5
|
| [29] |
KO SR, SUZUKI Y, SUZUKI K, CHOI KJ, CHO BG. Marked production of ginsenosides Rd, F2, Rg3, and compound K by enzymatic method. Chemical and Pharmaceutical Bulletin, 2007, 55(10): 1522-1527. DOI:10.1248/cpb.55.1522
|
| [30] |
YAN Q, ZHOU W, SHI XL, ZHOU P, JU DW, FENG MQ. Biotransformation pathways of ginsenoside Rb1 to compound K by β-glucosidases in fungus Paecilomyces bainier sp. 229. Process Biochemistry, 2010, 45(9): 1550-1556. DOI:10.1016/j.procbio.2010.06.007
|
| [31] |
CHANG KH, JO MN, KIM KT, et al. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. Journal of Ginseng Research, 2014, 38(1): 47-51. DOI:10.1016/j.jgr.2013.11.008
|
| [32] |
UPADHYAYA J, KIM MJ, KIM YH, KO SR, PARK HW, KIM MK. Enzymatic formation of compound-K from ginsenoside Rb1 by enzyme preparation from cultured mycelia of Armillaria mellea. Journal of Ginseng Research, 2016, 40(2): 105-112. DOI:10.1016/j.jgr.2015.05.007
|
| [33] |
KIM MJ, UPADHYAYA J, YOON MS, RYU NS, SONG YE, PARK HW, KIM YH, KIM MK. Highly regioselective biotransformation of ginsenoside Rb2 into compound Y and compound K by β-glycosidase purified from Armillaria mellea mycelia. Journal of Ginseng Research, 2018, 42(4): 504-511. DOI:10.1016/j.jgr.2017.07.001
|
| [34] |
KIM SA, JEONG EB, OH DK. Complete bioconversion of protopanaxadiol-type ginsenosides to compound K by extracellular enzymes from the isolated strain Aspergillus tubingensis. Journal of Agricultural and Food Chemistry, 2021, 69(1): 315-324. DOI:10.1021/acs.jafc.0c07424
|
| [35] |
QUAN LH, KIM YJ, LI GH, CHOI KT, YANG DC. Microbial transformation of ginsenoside Rb1 to compound K by Lactobacillus paralimentarius. World Journal of Microbiology and Biotechnology, 2013, 29(6): 1001-1007. DOI:10.1007/s11274-013-1260-1
|
| [36] |
KIM SY, LEE HN, HONG SJ, KANG HJ, CHO JY, KIM D, AMEER K, KIM YM. Enhanced biotransformation of the minor ginsenosides in red ginseng extract by Penicillium Decumbens β-glucosidase. Enzyme and Microbial Technology, 2022, 153: 109941. DOI:10.1016/j.enzmictec.2021.109941
|
| [37] |
KIM JK, CHOI MS, JEUNG W, RA J, YOO HH, KIM DH. Effects of gut microbiota on the pharmacokinetics of protopanaxadiol ginsenosides Rd, Rg3, F2, and compound K in healthy volunteers treated orally with red ginseng. Journal of Ginseng Research, 2020, 44(4): 611-618. DOI:10.1016/j.jgr.2019.05.012
|
| [38] |
DOU TY, CHEN J, QIAN XK, LI K, GE GB. Biotransformation of glycoginsenosides to intermediate products and aglycones using a hemicellulosome produced by Cellulosimicrobium cellulan. Applied Biochemistry and Microbiology, 2019, 55(2): 117-122. DOI:10.1134/S0003683819020054
|
| [39] |
HAN SK, JOO MK, KIM JK, JEUNG W, KANG H, KIM DH. Bifidobacteria-fermented red ginseng and its constituents ginsenoside Rd and protopanaxatriol alleviate anxiety/depression in mice by the amelioration of gut dysbiosis. Nutrients, 2020, 12(4): 901. DOI:10.3390/nu12040901
|
| [40] |
QUAN LH, JIN Y, WANG C, MIN JW, KIM YJ, YANG DC. Enzymatic transformation of the major ginsenoside Rb2 to minor compound Y and compound K by a ginsenoside-hydrolyzing β-glycosidase from Microbacterium esteraromaticum. Journal of Industrial Microbiology & Biotechnology, 2012, 39(10): 1557-1562.
|
| [41] |
ZHONG FL, MA R, JIANG ML, DONG WW, JIANG J, WU SQ, LI DH, QUAN LH. Cloning and characterization of ginsenoside-hydrolyzing β-glucosidase from Lactobacillus brevis that transforms ginsenosides Rb1 and F2 into ginsenoside Rd and compound K. Journal of Microbiology and Biotechnology, 2016, 26(10): 1661-1667. DOI:10.4014/jmb.1605.05052
|
| [42] |
YOO MH, YEOM SJ, PARK CS, LEE KW, OH DK. Production of aglycon protopanaxadiol via compound K by a thermostable β-glycosidase from Pyrococcus furiosus. Applied Microbiology and Biotechnology, 2011, 89(4): 1019-1028. DOI:10.1007/s00253-010-2960-1
|
| [43] |
SHIN KC, KIM TH, CHOI JH, OH DK. Complete biotransformation of protopanaxadiol-type ginsenosides to 20-O-β-glucopyranosyl-20 (S)-protopanaxadiol using a novel and thermostable β-glucosidase. Journal of Agricultural and Food Chemistry, 2018, 66(11): 2822-2829. DOI:10.1021/acs.jafc.7b06108
|
| [44] |
XIE JC, ZHAO DX, ZHAO LG, PEI JJ, XIAO W, DING G, WANG ZZ. Overexpression and characterization of a Ca2+ activated thermostable β-glucosidase with high ginsenoside Rb1 to ginsenoside 20 (S)-Rg3 bioconversion productivity. Journal of Industrial Microbiology & Biotechnology, 2015, 42(6): 839-850.
|
| [45] |
GERALDI A, NI'MATUZAHROH, FATIMAH, CUI CH, NGUYEN TT, KIM SC. Enzymatic biotransformation of ginsenoside Rb1 by recombinant β-glucosidase of bacterial isolates from Indonesia. Biocatalysis and Agricultural Biotechnology, 2020, 23: 101449. DOI:10.1016/j.bcab.2019.101449
|
| [46] |
SIDDIQI MZ, MEDJEBOURI S, LIU QZ, PARK HY, KIM GR, IM WT. Efficient production of various minor ginsenosides from PPD- and PPT-type major ginsenosides using a single recombinant BglFc isolated from Flavobacterium chilense. Biotechnology and Bioprocess Engineering, 2021, 26(2): 232-246. DOI:10.1007/s12257-020-0099-1
|
| [47] |
MÉNDEZ-LÍTER JA, NIETO-DOMÍNGUEZ M, FERNÁNDEZ de TORO B, GONZÁLEZ SANTANA A, PRIETO A, ASENSIO JL, CAÑADA FJ, de EUGENIO LI, MARTÍNEZ MJ. A glucotolerant β-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds. Microbial Cell Factories, 2020, 19(1): 127. DOI:10.1186/s12934-020-01386-1
|
| [48] |
CHENG LY, ZHANG H, CUI HY, DAVARI MD, WEI B, WANG WY, YUAN QP. Efficient enzyme-catalyzed production of diosgenin: inspired by the biotransformation mechanisms of steroid saponins in Talaromyces stollii CLY-6. Green Chemistry, 2021, 23(16): 5896-5910. DOI:10.1039/D0GC04152A
|
| [49] |
CAO LG, WU H, ZHANG H, ZHAO Q, YIN X, ZHENG DR, LI CW, KIM MJ, KIM P, XUE ZY, WANG Y, LI YH. Highly efficient production of diverse rare ginsenosides using combinatorial biotechnology. Biotechnology and Bioengineering, 2020, 117(6): 1615-1627. DOI:10.1002/bit.27325
|
| [50] |
LU Y, ZHOU JL, GU QY, YANG WH, YANG L, YU XB. Cloning, expression and improvement of catalytic activity of alginate lyase by site-directed mutation. Systems Microbiology and Biomanufacturing, 2022, 2(3): 555-567. DOI:10.1007/s43393-022-00084-w
|
| [51] |
SHIN KC, HONG SH, SEO MJ, OH DK. An amino acid at position 512 in β-glucosidase from Clavibacter michiganensis determines the regioselectivity for hydrolyzing gypenoside XVII. Applied Microbiology and Biotechnology, 2015, 99(19): 7987-7996. DOI:10.1007/s00253-015-6549-6
|
| [52] |
WILTSCHI B, CERNAVA T, DENNIG A, GALINDO CASAS M, GEIER M, GRUBER S, HABERBAUER M, HEIDINGER P, HERRERO ACERO E, KRATZER R, LULEY-GOEDL C, MÜLLER CA, PITZER J, RIBITSCH D, SAUER M, SCHMÖLZER K, SCHNITZHOFER W, SENSEN CW, SOH J, STEINER K, WINKLER CK, et al. Enzymes revolutionize the bioproduction of value-added compounds: from enzyme discovery to special applications. Biotechnology Advances, 2020, 40: 107520. DOI:10.1016/j.biotechadv.2020.107520
|
| [53] |
BARBOSA O, ORTIZ C, BERENGUER-MURCIA Á, TORRES R, RODRIGUES RC, FERNANDEZ-LAFUENTE R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnology Advances, 2015, 33(5): 435-456. DOI:10.1016/j.biotechadv.2015.03.006
|
| [54] |
SHI XJ, XU J, LU CN, WANG ZZ, XIAO W, ZHAO LG. Immobilization of high temperature-resistant GH3 β-glucosidase on a magnetic particle Fe3O4-SiO2-NH2-Cellu-ZIF8/zeolitic imidazolate framework. Enzyme and Microbial Technology, 2019, 129: 109347. DOI:10.1016/j.enzmictec.2019.05.004
|
| [55] |
KAZAN A, HU XH, STAHL A, FRERICHS H, SMIRNOVA I, YESIL-CELIKTAS O. An enzyme immobilized microreactor for continuous-flow biocatalysis of ginsenoside Rb1. Journal of Chemical Technology & Biotechnology, 2021, 96(12): 3349-3357.
|
| [56] |
LI WN, ZHANG XW, XUE ZY, MI Y, MA P, FAN DD. Ginsenoside CK production by commercial snailase immobilized onto carboxylated chitosan-coated magnetic nanoparticles. Biochemical Engineering Journal, 2021, 174: 108119. DOI:10.1016/j.bej.2021.108119
|
| [57] |
WU XH, QU BQ, LIU YK, REN XX, WANG SY, QUAN YL. Highly enhanced activity and stability via affinity induced immobilization β-glucosidase from Aspergillus niger onto amino-based silica for the biotransformation of ginsenoside Rb1. Journal of Chromatography A, 2021, 1653: 462388.
|
| [58] |
CHAMOLI S, YADAV E, Hemansi, SAINI JK, VERMA AK, NAVANI NK, KUMAR P. Magnetically recyclable catalytic nanoparticles grafted with Bacillus subtilis β-glucosidase for efficient cellobiose hydrolysis. International Journal of Biological Macromolecules, 2020, S0141-8130(20): 34190.
|
| [59] |
CHENG XQ, ZHOU XR, ZHENG ZP, KUANG Q. Construct efficient substrate transport and catalytic sub-nanochannels in metal-organic framework-based nanozymes for boosting peroxidase-like catalytic activity. Chemical Engineering Journal, 2022, 430: 133079. DOI:10.1016/j.cej.2021.133079
|
| [60] |
HAN J, FENG H, WU JC, LI YY, ZHOU Y, WANG L, LUO P, WANG Y. Construction of multienzyme Co-immobilized hybrid nanoflowers for an efficient conversion of cellulose into glucose in a cascade reaction. Journal of Agricultural and Food Chemistry, 2021, 69(28): 7910-7921. DOI:10.1021/acs.jafc.1c02056
|
| [61] |
LONG LF, HU Y, XIE L, SUN FB, XU ZH, HU JG. Constructing a bacterial cellulose-based bacterial sensor platform by enhancing cell affinity via a surface-exposed carbohydrate binding module. Green Chemistry, 2021, 23(23): 9600-9609. DOI:10.1039/D1GC03097C
|
| [62] |
LADOLE MR, POKALE PB, PATIL SS, BELOKAR PG, PANDIT AB. Laccase immobilized peroxidase mimicking magnetic metal organic frameworks for industrial dye degradation. Bioresource Technology, 2020, 317: 124035. DOI:10.1016/j.biortech.2020.124035
|
| [63] |
STEPANKOVA V, BIDMANOVA S, KOUDELAKOVA T, PROKOP Z, CHALOUPKOVA R, DAMBORSKY J. Strategies for stabilization of enzymes in organic solvents. ACS Catalysis, 2013, 3(12): 2823-2836. DOI:10.1021/cs400684x
|
| [64] |
ZHANG QH, de OLIVEIRA VIGIER K, ROYER S, JÉRÔME F. Deep eutectic solvents: syntheses, properties and applications. Chemical Society Reviews, 2012, 41(21): 7108-7146. DOI:10.1039/c2cs35178a
|
| [65] |
BI YH, ZHU C, WANG ZY, LUO HZ, FU RP, ZHAO XJ, ZHAO XJ, JIANG L. Purification and characterization of a glucose-tolerant β-glucosidase from black plum seed and its structural changes in ionic liquids. Food Chemistry, 2019, 274: 422-428. DOI:10.1016/j.foodchem.2018.09.007
|
| [66] |
CAÑADAS R, GONZÁLEZ-MIQUEL M, GONZÁLEZ EJ, DÍAZ I, RODRÍGUEZ M. Overview of neoteric solvents as extractants in food industry: a focus on phenolic compounds separation from liquid streams. Food Research International (Ottawa, Ont), 2020, 136: 109558. DOI:10.1016/j.foodres.2020.109558
|
| [67] |
GULLÓN P, GULLÓN B, ROMANÍ A, ROCCHETTI G, LORENZO JM. Smart advanced solvents for bioactive compounds recovery from agri-food by-products: a review. Trends in Food Science & Technology, 2020, 101: 182-197.
|
| [68] |
LIU Y, ZHENG JY, XIAO JX, HE XD, ZHANG KX, YUAN SX, PENG ZT, CHEN Z, LIN XQ. Enhanced enzymatic hydrolysis and lignin extraction of wheat straw by triethylbenzyl ammonium chloride/lactic acid-based deep eutectic solvent pretreatment. ACS Omega, 2019, 4(22): 19829-19839. DOI:10.1021/acsomega.9b02709
|
| [69] |
ZOU YC, XIN X, XU HX, YUAN HW, LI XF, YU YG, ZHAO GL. Highly efficient bioconversion of flavonoid glycosides from citrus-processing wastes in solvent-buffer systems. Green Chemistry, 2020, 22(10): 3196-3207. DOI:10.1039/D0GC00669F
|
| [70] |
JEONG KM, LEE MS, NAM MW, ZHAO J, JIN Y, LEE DK, KWON SW, JEONG JH, LEE J. Tailoring and recycling of deep eutectic solvents as sustainable and efficient extraction media. Journal of Chromatography A, 2015, 1424: 10-17.
|
| [71] |
LI P, ZHAO PY, LIU WJ, JIANG YF, WANG WJ, BAO LY, JIN YR, LI XW. Determination of common ginsenosides in Kang'ai injection by aqueous two-phase extraction with deep eutectic solvents and HPLC-UV/DAD. Microchemical Journal, 2018, 137: 302-308. DOI:10.1016/j.microc.2017.11.007
|
| [72] |
HAN X, LI WN, MA XX, FAN DD. Enzymatic hydrolysis and extraction of ginsenoside recovered from deep eutectic solvent-salt aqueous two-phase system. Journal of Bioscience and Bioengineering, 2020, 130(4): 390-396. DOI:10.1016/j.jbiosc.2020.05.008
|
| [73] |
MA Z, MI Y, HAN X, LI HH, TIAN M, DUAN ZG, FAN DD, MA P. Transformation of ginsenoside via deep eutectic solvents based on choline chloride as an enzymatic reaction medium. Bioprocess and Biosystems Engineering, 2020, 43(7): 1195-1208. DOI:10.1007/s00449-020-02314-8
|
| [74] |
SMITH EL, ABBOTT AP, RYDER KS. Deep eutectic solvents (DESs) and their applications. Chemical Reviews, 2014, 114(21): 11060-11082. DOI:10.1021/cr300162p
|
| [75] |
WANG DQ, ZHENG P, CHEN PC, WU D. Highly efficient enzymatic conversion of rutin to isoquercitrin and l-rhamnose using deep eutectic solvents. ACS Sustainable Chemistry & Engineering, 2020, 8(39): 14905-14913.
|
| [76] |
CHENG QB, ZHANG LW. Highly efficient enzymatic preparation of daidzein in deep eutectic solvents. Molecules (Basel, Switzerland), 2017, 22(1): 186. DOI:10.3390/molecules22010186
|
| [77] |
XU KJ, WANG YZ, HUANG YH, LI N, WEN Q. A green deep eutectic solvent-based aqueous two-phase system for protein extracting. Analytica Chimica Acta, 2015, 864: 9-20. DOI:10.1016/j.aca.2015.01.026
|
| [78] |
HOPPE J, BYZIA E, SZYMAŃSKA M, DROZD R, SMIGLAK M. Acceleration of lactose hydrolysis using beta-galactosidase and deep eutectic solvents. Food Chemistry, 2022, 384: 132498. DOI:10.1016/j.foodchem.2022.132498
|
| [79] |
NI JC, LI Q, GONG L, LIAO XL, ZHANG ZJ, MA CL, HE YC. Highly efficient chemoenzymatic cascade catalysis of biomass into furfurylamine by a heterogeneous shrimp shell-based chemocatalyst and an ω-transaminase biocatalyst in deep eutectic solvent-water. ACS Sustainable Chemistry & Engineering, 2021, 9(38): 13084-13095.
|
| [80] |
LOMBA L, GARCÍA CB, RIBATE MP, GINER B, ZURIAGA E. Applications of deep eutectic solvents related to health, synthesis, and extraction of natural based chemicals. Applied Sciences, 2021, 11(21): 10156. DOI:10.3390/app112110156
|
| [81] |
WANG PP, WEI YJ, FAN Y, LIU QF, WEI W, YANG CS, ZHANG L, ZHAO GP, YUE JM, YAN X, ZHOU ZH. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metabolic Engineering, 2015, 29: 97-105. DOI:10.1016/j.ymben.2015.03.003
|
| [82] |
KIM YJ, ZHANG DB, YANG DC. Biosynthesis and biotechnological production of ginsenosides. Biotechnology Advances, 2015, 33(6): 717-735. DOI:10.1016/j.biotechadv.2015.03.001
|
| [83] |
WANG PP, WANG JL, ZHAO GP, YAN X, ZHOU ZH. Systematic optimization of the yeast cell factory for sustainable and high efficiency production of bioactive ginsenoside compound K. Synthetic and Systems Biotechnology, 2021, 6(2): 69-76. DOI:10.1016/j.synbio.2021.03.002
|
 2023, Vol. 39
2023, Vol. 39




