中国科学院微生物研究所、中国微生物学会主办
文章信息
- 鲍江舰, 杨君仪, 邵瑞瑞, 张婷, 廖健, 程玉梅, 官志忠, 齐晓岚, 陈峥宏, 洪伟, 崔古贞
- BAO Jiangjian, YANG Junyi, SHAO Ruirui, ZHANG Ting, LIAO Jian, CHENG Yumei, GUAN Zhizhong, QI Xiaolan, CHEN Zhenghong, HONG Wei, CUI Guzhen
- fliL基因显著影响艰难拟梭菌运动功能及产孢能力
- The fliL gene significantly affects the motility and sporulation abilities of Clostridioides difficile
- 生物工程学报, 2023, 39(4): 1578-1595
- Chinese Journal of Biotechnology, 2023, 39(4): 1578-1595
- 10.13345/j.cjb.220765
-
文章历史
- Received: September 24, 2022
- Accepted: December 6, 2022
- Published: December 9, 2022
2. 贵州医科大学 地方病与少数民族疾病教育部重点实验室 贵州省医学分子生物学重点实验室, 贵州 贵阳 550001;
3. 省部共建地方病及民族区域性疾病防控协同创新中心, 贵州 贵阳 550001;
4. 贵州医科大学口腔医学院 附属口腔医院, 贵州 贵阳 550001;
5. 贵州医科大学附属医院综合ICU, 贵州 贵阳 550001;
6. 贵州医科大学病理科 贵州医科大学附属医院, 贵州 贵阳 550001
2. Key Laboratory of Medical Molecular Biology of Guizhou Province, Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education, Guizhou Medical University, Guiyang 550001, Guizhou, China;
3. Collaborative Innovation Center for Prevention and Control of Endemic and Ethnic Regional Diseases Co-Constructed by the Province and Ministry, Guiyang 550001, Guizhou, China;
4. Affiliated Stomatological Hospital, School of Stomatology, Guizhou Medical University, Guiyang 550001, Guizhou, China;
5. Department of Critical Care Medicine, the Affiliated Hospital of Guizhou Medical University, Guiyang 550001, Guizhou, China;
6. The Affiliated Hospital of Guizhou Medical University, Department of Pathology, Guizhou Medical University, Guiyang 550001, Guizhou, China
艰难拟梭菌是一种严格厌氧、革兰氏阳性产芽孢杆菌[1]。人类肠道正常菌群可以有效地抑制艰难拟梭菌的定殖,但抗菌药物及免疫抑制剂的使用会引起肠道菌群紊乱,导致艰难拟梭菌感染(Clostridioides difficile infection, CDI)。CDI可引起腹泻、肠出血、肠穿孔及中毒性巨结肠等症状。近20年以来,随着高毒力NAP1/ BI/027菌株的出现和传播,医院获得性CDI (hospital acquired CDI, HA-CDI)的病人成倍增长。2000年至2017年北美国家HA-CDI导致病人死亡比例由小于1.5%增长至4.5%−5.7%,在流行期间甚至达到16.7%。在我国,CDI的发病率及复发率亦逐年提升[1-4]。艰难拟梭菌引起的腹泻已逐渐成为抗生素相关性腹泻的主要类型。
鞭毛作为艰难拟梭菌主要的运动结构,其介导的运动能力影响着艰难拟梭菌在人体胃肠道的粘附、定殖及其毒力。鞭毛由鞭毛基体、鞭毛钩及鞭毛丝组成,鞭毛基体中包含多个定子单元及一个转子以实现细菌的运动功能。鞭毛基体的转子是由作为中央驱动轴的杆部及各类环状功能结构组成,定子则是作为“能量转换器”将离子通道的跨膜运输能量转换为力矩促进细菌运动[5]。fliL基因编码的FliL蛋白是一种与鞭毛基体相结合的单跨膜蛋白[6]。有研究通过断层分析表明在伯氏疏螺旋体中FliL蛋白位于转子及定子附近[7]。大量研究成果表明FliL蛋白在不同的菌株中发挥的作用并不完全相同,FliL蛋白对于新月柄杆菌(Caulobacter crescentus)和类球红细菌(Rhodobacter sphaeroides)的运动是必需的[8-9]。Attmannspacher等报道大肠杆菌和沙门氏菌fliL基因突变株在游泳运动能力上略有降低[10]。Chawla等在后续的研究中甚至得出了相反的结论即fliL基因并不是大肠杆菌运动所必需的结构[11]。可见,FliL蛋白的缺失在不同菌株中导致的表型变化并不一致。艰难拟梭菌fliL基因对其表型及致病力的影响目前尚不明确,仍需进一步研究。
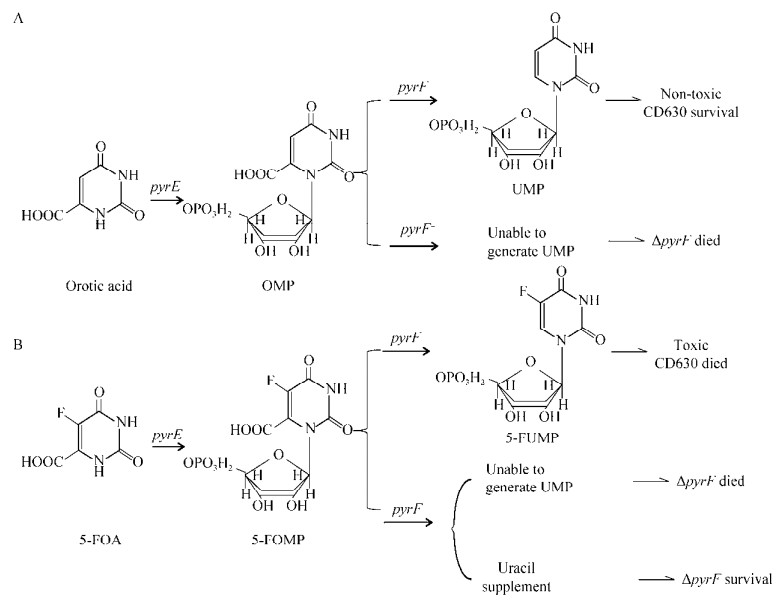
目前各种不同的基因编辑技术被应用于艰难拟梭菌的遗传改造[12-14]。接合转化在质粒转化时具有避开胞外核酸酶的显著优势,目前接合转化已大量用于艰难拟梭菌的基因编辑,其主要通过性菌毛将遗传物质从供体菌转移给受体菌。DNA转移至艰难拟梭菌并在菌株内进行稳定复制一直是限制艰难拟梭菌相关研究的难题。2002年Purdy等在艰难拟梭菌CD630中首次发现PCD6质粒,并将促进了该质粒复制的主要功能元件(orfA)成功分离。以orfA为基础构建的pMTL9301质粒成功地通过接合转化从大肠杆菌CA434菌株转移至艰难拟梭菌中并进行稳定复制[15],至此完善了穿梭质粒接合转化至艰难拟梭菌的技术。众多编辑技术中基于尿嘧啶缺陷菌株ΔpyrE的非等长同源臂偶联等位交换(allele-coupled exchange, ACE)编辑方法由Heap等建立,并成功应用于艰难拟梭菌中揭示了细胞壁结合半胱氨酸蛋白酶(cell wall-binding cysteine protease, Cwp84)的功能[16-17]。其原理在于乳清酸磷酸核糖基转移酶(orotate phosphoribosyl transferase, pyrE)基因缺失时艰难拟梭菌无法从头合成尿嘧啶导致细胞死亡,相反在该基因存在时细菌可以将5-氟乳清酸(5-fluoroorotic acid, 5-FOA)转化成具有毒性的5-氟尿嘧啶(5-fluorouracil, 5-FU)导致菌体死亡[18](图 1)。另一方面ACE技术通过非等长同源臂等位交换原理可控制同源重组发生顺序,实现对目标基因的可控敲除[19]。
|
| 图 1 尿嘧啶从头合成途径 Fig. 1 Partial steps of the pyrimidine de novo biosynthesis pathway. A: Orotic acid was converted to non-toxic UMP by orotate phosphoribosyl transferase (pyrE) and orotidine-5′-phosphate decarboxylase (pyrF). B: 5-fluoroorotic acid (5-FOA) was converted to toxic 5-fluorouracil (5-FUMP) by pyrE and pyrF. |
| |
本研究中,我们建立了尿嘧啶从头合成过程中的必需基因(orotidine-5′-phosphate decarboxylase, pyrF)的缺陷菌株ΔpyrF (图 1),以该菌株作为底盘细胞,使用非等长同源臂偶联等位交换(allele-coupled exchange, ACE)方法构建鞭毛基底体相关FliL家族蛋白(flagellar basal body-associated FliL family protein, fliL)基因缺失菌株和回补菌株[16]。通过对比野生型菌株、∆fliL及其回补突变株在生长曲线、运动能力、抗生素敏感性及产孢能力等表型变化,研究艰难拟梭菌FliL蛋白生理、生化功能及其对艰难拟梭菌致病力的影响。
1 材料与方法 1.1 菌株和培养条件本研究中使用的菌株及质粒信息均列于表 1。选用大肠杆菌(Escherichia coli) (NEB Express Competent)作为宿主,通过化学转化的方式完成质粒DNA构建。以大肠杆菌CA434作为宿主,通过接合转化将大肠杆菌CA434的质粒DNA转移至艰难拟梭菌菌株CD630[19]。大肠杆菌菌株NEB Express Competent及大肠杆菌菌株CA434在添加氯霉素(25 μg/mL)的Luria-Bertani (LB)培养基中37 ℃培养。LB培养基配方:1%胰蛋白胨,0.5%酵母提取物,1%氯化钠。艰难拟梭菌CD630生长在脑心浸出液补充(brain heart infusion supplemented, BHIS)培养基中培养(37 ℃,厌氧环境),BHIS培养基配方为:37.5% BHI培养基,0.5%半胱氨酸盐酸盐,1%酵母提取物。培养基中加入甲砜霉素(15 μg/mL)、头孢西丁(16 μg/mL)、D环丝氨酸(125 μg/mL)筛选艰难拟梭菌转化子。5-FOA和尿嘧啶购买自北京索莱宝科技有限公司,5-FOA (100 mg/mL)被溶解在二甲基亚砜(dimethyl sulfoxide, DMSO)中,尿嘧啶(20 mg/mL)被溶解在双蒸水中。筛选ΔpyrF菌株及ΔpyrFΔfliL时5-FOA及尿嘧啶被加入到BHIS培养基或艰难拟梭菌基本培养基(Clostridioides difficile minimal medium, CDMM)培养基中[19]。琼脂粉(1.5%)加入LB培养基、BHIS培养基和CDMM培养基中制备固体培养基。
| Strains and plasmid | Features | Resources |
| NEB express competent E. coli (high efficiency) |
General cloning host for plasmid manipulation | NEB |
| CA434 | Donor strain for conjugation between E. coli and Streptomyces | Lab stock |
| CD630 | Wild-type Clostridioides difficile 630 strain (CD630) | American Type Culture Collection, ATCC |
| ΔpyrF | pyrF gene disruption mutant, derived from CD630 | This work |
| ΔpyrFΔfliL | pyrF gene and fliL gene disruption mutant, derived from ΔpyrF | This work |
| ΔfliL | pyrF gene complemented strain, derived from ΔpyrFΔfliL | This work |
| : : fliL | pyrF and fliL complemented strain, derived from ΔpyrFΔfliL | This work |
| pMTL82151 | pBP1 ori, CmR, ColE1 ori, traJ, lacZ α fragment | [a] |
| pMTL-BY | For deletion of the pyrF gene, containing upstream and downstream homologous arms of pyrF, derived from pMTL82151 |
This work |
| pMTL-BJJ | Derived from pMTL82151, for deletion of the fliL gene, containing upstream and downstream homologous arms of fliL |
This work |
| pMTL-BJ1 | Derived from pMTL-BJJ, containing pyrF gene and promoter from Clostridium beijerinckii NCIMB 8052 |
This work |
| pMTL-BJ2 | Derived from pMTL82151, for complement for pyrF gene, containing CD630_pyrF gene and promoter |
This work |
| pMTL-BJ3 | Derived from pMTL-BJ2, for complement for fliL gene, containing fliL gene and promoter |
This work |
| [a]: Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. Journal of Microbiological Methods 78: 79–85. | ||
本研究中所使用的引物信息列于表 2。DNA限制性内切酶购自NEB公司,DNA聚合酶(2× Phanta Flash Master Mix和2× Rapid Taq Master Mix)购自南京诺唯赞生物科技股份有限公司。
| Primer | Primer sequence (5′→3′) | Usages |
| HW731 | GCCTGCAGACATGCAAGCATGCAAACAGTGCAAAAAAT | Amplification of CD630_pyrF deletion fragment |
| HW732 | GTTATAGGTCTTCCAACAACAGCTTTATCAAATTCGTCAGT | |
| HW733 | ACTGACGAATTTGATAAAGCTGTTGTTGGAAGACCTATAACAA | |
| HW734 | ACGACGGCCAGTGCCAAGCCCTGGGCCATTAGATAATAAG | |
| HW735 | ACGACGGCCAGTGCCAAGCTATTGGTCCTGACCCTAATAC | Detection of CD630_pyrF |
| HW736 | AGACTGGGAAAGATATCTCAT | |
| HW681 | ATCAGGAAACAGCTATGACCG | Detection of pMTL82151 multiple cloning sites |
| HW682 | GTTTTCCCAGTCACGACGTT | |
| HW601 | TTTTTTGTTACCCTAAGTTTTAAAACTAGACACTAATGATTGC | Amplify 8052_pyrF |
| HW602 | AGATTATCAAAAAGGAGTTTTTATATGTTCTTCACTGCTTCT | |
| HW605 | ATCGTAGAAATACGGTGTTT | Detection of pyrF in pMTL-BJJ |
| HW606 | ACGTTAAGGGATTTTGGTCA | |
| HW639 | CGAGGCCTGCAGACATGCAATATTTAGTACGGGTTCAAGT | Amplification of the fliL deletion fragment |
| HW640 | GTATCATGTCTTACCATCCTAAACCTCTTCCTCCTAGTCA | |
| HW641 | TGACTAGGAGGAAGAGGTTTAGGATGGTAAGACATGATAC | |
| HW642 | CGACGGCCAGTGCCAAGCTAGTATGTAACATCCAGCTTC | |
| HW797 | TAAAAACATAGCAGATGCGT | Detection of fliL |
| HW798 | GTGTTCCAAAGAATGCTACTA | |
| HW821 | GTCACGCGTCCATGGAGATCAACATAGCAGATGCGTTAAA | Amplify fliL complement fragment |
| HW822 | GCTTGCATGTCTGCAGGCCGTATCATGTCTTACCATCCT | |
| HW554 | GGGAGACTTGAGTGCAGGAG | Amplify rrs gene for RT-qPCR |
| HW555 | GTGCCTCAGCGTCAGTTACAGT | |
| HW875 | TGCAGGTTTGTTTATTCCAACAGG | Amplify fliL gene for RT-qPCR |
| HW876 | CACCTTCATCAGCTAGCTTTAATACCA |
本研究中采用引物HW731/HW732以CD630基因组为模板扩增CD630_pyrF基因上游同源臂(798 bp),采用引物HW733/HW734以CD630基因组为模板扩增CD630_pyrF基因下游同源臂(798 bp)。CD630_pyrF基因上下游片段产物为模板,采用HW731/HW734引物扩增缺失CD630_pyrF基因片段。使用ClonExpress MultiS One Step Cloning Kit同源重组试剂,将上述缺失CD630_pyrF基因的聚合酶链式反应(polymerase chain reaction, PCR)产物与pMTL82151质粒(通过Hind Ⅲ线性化)连接,构建pyrF敲除质粒pMTL-BY。采用HW681/ HW682验证质粒是否构建成功。
1.2.2 fliL基因缺失质粒构建以CD630基因组为模板,采用HW639/ HW640及HW641/HW642引物,扩增fliL基因缺失片段(1 846 bp)。pMTL-BJJ质粒通过同源重组连接fliL基因缺失片段及pMTL-82151质粒(通过Hind Ⅲ线性化)构建。引物HW681/ HW682验证pMTL-BJJ质粒(1 983 bp)是否构建成功。采用引物HW601/HW602以拜氏梭菌(Clostridium beijerinckii) NCIMB 8052菌株基因组为模板扩增其的pyrF基因及其启动子片段(2 875 bp),ClonExpress MultiS One Step Cloning Kit同源重组试剂将扩增的菌株8052的pyrF基因片段及Pme Ⅰ线性化pMTL-BJJ质粒连接。采用引物HW606/HW605验证构建完成的质粒pMTL-BJ1。
1.2.3 pyrF回补质粒构建构建pyrF回补质粒pMTL-BJ2时,采用引物HW731/HW734以CD630基因组为模板扩增完整CD630_pyrF基因及启动子片段,通过同源重组方法将扩增的CD630_pyrF基因及启动子片段及Hind Ⅲ线性化pMTL82151质粒连接。采用HW681/HW682对构建完成的质粒进行验证。
1.2.4 fliL基因回补质粒构建以CD630基因组为模板,采用HW821/ HW822引物,扩增fliL基因及其启动子片段(1 070 bp)。通过同源重组方法将fliL基因及其启动子片段及pMTL-BJ2 (Xho Ⅰ线性化)质粒连接构建pMTL-BJ3。
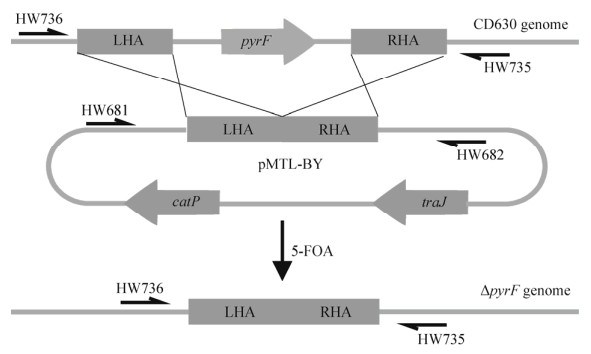
1.3 突变菌株构建 1.3.1 接合转化及pyrF基因敲除按照1.2中所述的方法构建pMTL-BY质粒。将质粒偶联到艰难梭菌中,以大肠杆菌CA434菌株作为质粒供体质粒偶联到艰难梭菌中,得到携带pMTL-BY质粒抗耐氯霉素的转化子。接合转化方法参考文献[20]。将上述转化子在添加了400 μg/mL 5-FOA的BHIS平板上培养48 h。待平板上发现菌落,采用HW735/ HW736引物鉴定pyrF基因是否敲除(图 2)。
|
| 图 2 同源重组方法构建ΔpyrF突变株 Fig. 2 Construction of ΔpyrF mutant by recombination method. Schematic diagram of the construction of ∆pyrF mutants. The RHA (right-side homology arm) and LHA (left-side homology arm) and on pMTL-BY plasmid recombinant with CD630 genome under 5-FOA stress. The colonies resistant to 5-FOA are screened by primer set HW736/HW735. |
| |
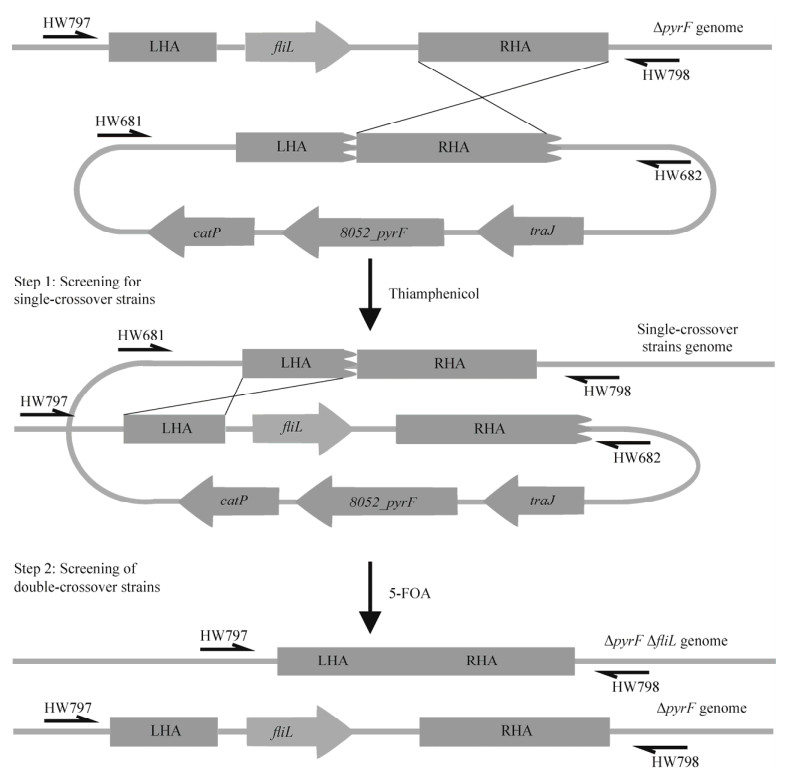
ΔpyrFΔfliL基因敲除突变株的构建过程,分为单交换菌株和双交换突变株的筛选(图 3)。首先将构建pMTL-BJ1质粒接合转化ΔpyrF菌株,在CDMM甲砜霉素(thiamphenicol, Tm)抗性平板(CDMM-Tm)培养基上选择耐Tm的转化子以获得单交换菌株。菌落是否为纯合单交换采用两对不同的引物进行PCR验证。第一对引物结合位点为pMTL-BJ1质粒LHA外侧(HW681)和CD630基因组RHA外侧(HW798),用于检测是否发生单交换;第二对引物结合位点为CD630基因组LHA外侧(HW797)和RHA外侧(HW798),用于检测是否存在野生型。如果第一对引物扩增产生特异性条带,且第二对引物不能扩增出条带,该菌落即为纯合单交换菌株。接下来,将获得的纯合单交换菌株涂布在含有2 mg/mL 5-FOA和20 μmol/mL尿嘧啶的CDMM培养基上,以筛选ΔpyrFΔfliL突变菌株,引物HW797/HW798用于检测fliL基因是否敲除(图 3)。
|
| 图 3 基于ACE方法构建突变株ΔpyrFΔfliL Fig. 3 Construction of the ΔpyrFΔfliL mutant strain by the ACE method. Schematic diagram of the construction of single-crossover and double-crossover mutants. The longer RHA (right-side homology arm) on the pMTL-BJJ plasmid has a higher recombination rate and tends to exchange with the ∆pyrF genome at the RHA site to generate single-crossover mutants. The obtained single-crossover mutants were spread on the 5-FOA-containing BHIS medium to screen 5-FOA-resistant colonies. The LHA (left-side homology arm) of single-crossover mutants recombined with the LHA on the genome to generate double-crossover mutants (ΔpyrFΔfliL). The primers pair HW797/HW798 was used to screen double-crossover mutants. |
| |
根据1.2中描述的方法构建回补载体pMTL-BJ2和pMTL-BJ3。回补载体pMTL-BJ2和pMTL-BJ3接合转化菌株ΔpyrFΔfliL,于BHIS甲砜霉素、d-环丝氨酸及头孢西丁抗性平板(BHIS-TDC)筛选后得到菌株ΔfliL和: : fliL。
1.4 定量逆转录PCR (quantitative reverse transcription PCR, RT-qPCR)验证fliL基因表达使用细菌总RNA提取试剂盒[天根生化科技(北京)有限公司]分别提取CD630、ΔfliL、: : fliL菌株总RNA。采用FasKing gDNA Dispelling RT SuperMix [天根生化科技(北京)有限公司]一步法去除基因组DNA并将总RNA逆转录成cDNA。以16S rRNA基因为内参基因对fliL基因表达量进行分析。使用Prism 6.0软件(GraphPad Software, Inc)绘制柱状图。
1.5 突变菌株生长速率测定将1% (体积分数)的艰难拟梭菌菌株(WT、ΔfliL、: : fliL)接种到5 mL BHIS培养基中。将上述接种菌株置于37 ℃厌氧培养。每隔2 h通过细胞密度计(Ultrospec 10, Amersham Biosciences, GE)测量OD600值,持续28 h。使用Prism 6.0软件(GraphPad Software, Inc)绘制生长曲线。
1.6 运动能力测定野生型CD630、ΔfliL及: : fliL在0.3%低浓度琼脂的半固体培养基上测定运动能力,方法参考文献[21]。观察菌株向培养基四周游动的距离。菌株向培养基四周扩散距离越远,表示该菌的运动越强,反之则表示该菌的运动能力越弱。
1.7 抗生素敏感性为了测定CD630、ΔfliL及: : fliL菌株对于不同抗生素的敏感性。采用连续稀释法于96孔板中设置不同的抗生素浓度梯度以测定CD630、ΔfliL、: : fliL菌株对于氨苄青霉素(ampicillin)、四环素(tetracycline)、阿莫西林(amoxicillin)、诺氟沙星(norfloxacin)、链霉素(streptomycin)、D环丝氨酸(d-cycloserine)、万古霉素(vancomycin)、克林霉素(clindamycin)、卡那霉素(kanamycin)等抗生素的耐药性。测定培养物OD600值并使用Prism 6.0软件(GraphPad Software, Inc)绘制柱状图。
1.8 pH耐受性测定将野生型CD630菌株、ΔfliL及: : fliL培养至OD600=0.6,细菌培养物分别取10 μL加入96孔板的BHIS培养基中,BHIS培养基已调整pH值为:1、2、3、4、5、6、7、8、9、10、11、12。菌株在厌氧工作站中培养24 h,观察各个pH值条件下不同菌株的生长情况。测定培养物OD600值并使用Prism 6.0软件(GraphPad Software, Inc)绘制柱状图。
1.9 产孢能力收集野生型CD630、ΔfliL及: : fliL菌株的孢子,方法参考文献[22]。收集的孢子通过PBS洗涤2次后置于60 ℃水浴30 min使菌株失活。孢子收集后分别取各个孢子20 μL接种于BHIS固体培养基并置于37 ℃厌氧培养5 d。孢子萌发后对平板上的菌落进行计数。
1.10 统计学方法本研究采用Prism 8软件及IBM SPSS Statistics 26软件进行统计学分析。每个实验采取3个平行重复,采用t检验分析组间差异,检测水平为P=0.05,*: P < 0.05,**: P < 0.01,***: P < 0.001,n表示P > 0.05,当P < 0.05时具有统计学意义。
2 结果与分析 2.1 ΔpyrF菌株构建及抗性分析我们成功构建了pMTL-BY质粒用于pyrF基因的敲除。pMTL-BY质粒构建的方法见方法1.2。根据上文所提及的方式成功地在含5-FOA的BHIS培养基上获得抗性艰难拟梭菌单菌落。通过引物HW735/HW736随机对培养基上生长的20个菌落进行PCR验证,20个菌落均为∆pyrF突变基因型(图 4A)。通过不断地对抗性菌株在无抗培养基中进行传代,ΔpyrF突变株中质粒成功丢失。将野生型菌株及构建的ΔpyrF同时接种于含5-FOA (2 mg/mL)的BHIS培养基培养48 h后,ΔpyrF菌株均可生长,而野生型的菌株生长均受到抑制,而接种在CDMM限制性培养基上的情况则相反。以上结果表明ΔpyrF突变株构建成功。
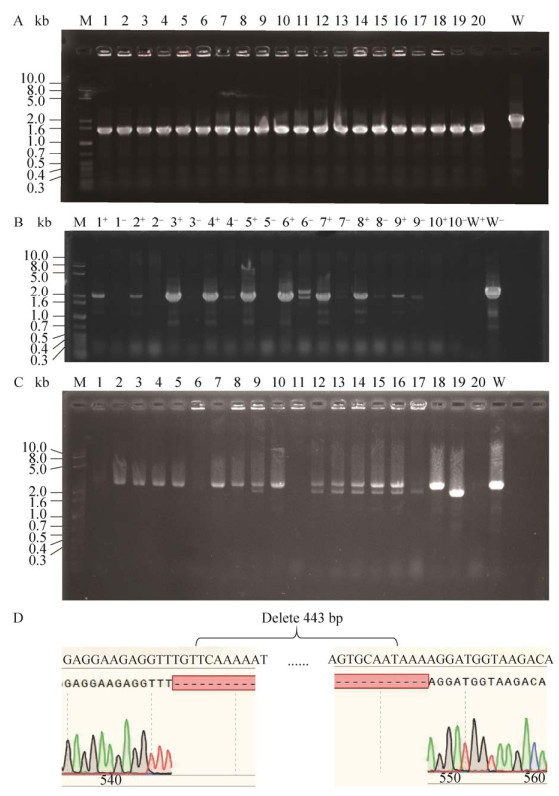
|
| 图 4 突变菌株筛选及打靶位点测序验证 Fig. 4 Mutants screening by PCR and verification by sequencing. A: All detected colonies (20) contained the ∆pyrF gene mutation (wild-type CD630 vs. ΔfliL=2 014 bp vs. 1 598 bp). M: 1 kb plus DNA marker; W: Wild-type CD630 strain. The gene targeting efficiency was 100%. B: Ten randomly selected colonies were detected by primer sets HW681/HW798 (+) and HW797/HW798 (–). Pure single-crossover mutants would produce a 2 046 bp band in the '+' lane and no band at the '–' lane. M: 1 kb plus DNA marker; W represents the wild-type CD630 strain that was used as the template. C: Twenty colonies were randomly selected and screened using the primer set HW797/HW798. M: 1 kb plus DNA marker. Two of the twenty strains showed ΔfliL bands compared to the wild-type control (wild-type CD630 vs. ΔfliL=2 405 bp vs. 1 982 bp) (W). D: Gene sequencing results showed that the fliL was deleted from the ΔpyrFΔfliL mutants. |
| |
以ΔpyrF菌株为底盘细胞,对目的基因fliL进行敲除。将pMTL-BJ1接合转化至ΔpyrF菌株,并通过菌落PCR对CDMM-Tm平板上的转化子进行验证。采用两对不同的引物进行PCR验证(方法1.3.2),筛选纯合的单交换菌株(图 4B)。因为双交换菌株从头合成尿嘧啶的代谢通路缺失(ΔpyrF),可以在BHIS (5-FOA 2 mg/mL)固体培养基上生存。故而将单交换菌株涂布于BHIS (5-FOA 2 mg/mL)固体培养基上筛选双交换菌株。采用引物HW797/HW798对5-FOA抗性菌落的fliL基因进行筛选与测序验证,最终获得ΔpyrFΔfliL双突变菌株(图 3,图 4C–D)。
2.3 回补菌株构建为了进一步验证fliL基因功能,我们分别构建了回补载体pMTL-BJ2和pMTL-BJ3 (表 1–2)。pMTL-BJ2具有pyrF基因及其启动子,在此基础上将fliL基因及其启动子重组至pMTL-BJ2母载体,获得pMTL-BJ3。从而将pyrF基因及fliL基因在质粒上进行表达。回补质粒pMTL-BJ2和pMTL-BJ3转化至ΔpyrFΔfliL后获得ΔfliL及: : fliL菌株。
2.4 fliL基因表达量变化通过RT-qPCR测定了野生型菌株CD630、菌株ΔfliL及: : fliL中fliL基因的表达情况。与CD630菌株相比较,ΔfliL菌株中fliL基因表达量显著降低;: : fliL菌株中fliL基因表达量显著提高(图 5)。
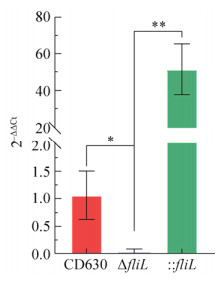
|
| 图 5 野生型菌株CD630、ΔfliL与: : fliL的fliL基因表达量比较 Fig. 5 Compare gene expression levels of the wild-type strain CD630, ΔfliL and : : fliL strains. The red bar represents the expression level of the fliL gene in the WT CD630 strain, the blue bar represents the gene expression level of fliL in the ΔfliL, the green bar represents the gene expression level of fliL in the strain : : fliL. *: P < 0.05; **: P < 0.01. |
| |
我们测定了野生型菌株和ΔfliL菌株的生长速率。与野生型CD630菌株相比较,ΔfliL菌株在前6 h生长速率无明显区别,在6 h后ΔfliL菌株生长速率低于CD630。CD630及ΔfliL菌株生长至平台期的时间也不相同,ΔfliL在生长13 h后进入平稳期,比11 h达到平稳期的CD630生长缓慢(图 6)。以上结果表明fliL突变株生长速率及其最大生物量(8−13 h)显著低于野生型菌株;fliL基因回补后,可见菌株的生长速率及最大生物量(8−13 h)恢复至野生型水平。
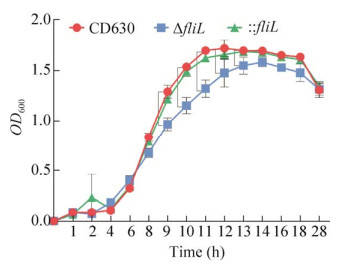
|
| 图 6 野生型CD630、ΔfliL与: : fliL生长曲线比较 Fig. 6 Comparison of growth profiles of strains CD630, ΔfliL and : : fliL. The growth profiles of CD630, ΔfliL and : : fliL mutants. The red circle represents the growth profile of strain CD630, The blue square represents the growth profile of strain ΔfliL, the green triangle represents the growth profile of strain : : fliL. |
| |
我们观察了菌株CD630、ΔfliL及: : fliL在0.3%浓度琼脂的半固体培养基上运动半径的变化。结果表明ΔfliL在0.3%浓度琼脂的半固体培养基上运动的范围显著小于野生型菌株CD630 (P < 0.000 1)。有趣的是: : fliL回补菌株的运动范围显著增大(P < 0.000 1) (图 7)。结果表明fliL基因是艰难拟梭菌实现游泳运动的关键基因。
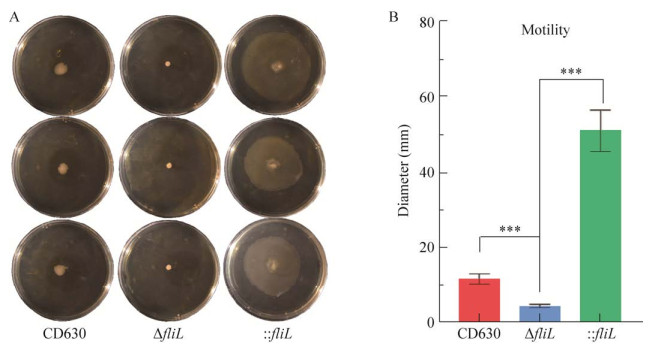
|
| 图 7 野生型CD630、ΔfliL与: : fliL的运动能力测定 Fig. 7 Motility of wild-type CD630, ΔfliL and : : fliL strains. A: The motility of strains CD630, ΔfliL and : : fliL on 0.3% agar BHIS medium. B: The statistical analysis of figure 6A. The swimming radius of ΔfliL was significantly smaller than that of CD630 and : : fliL. ***: P < 0.001. |
| |
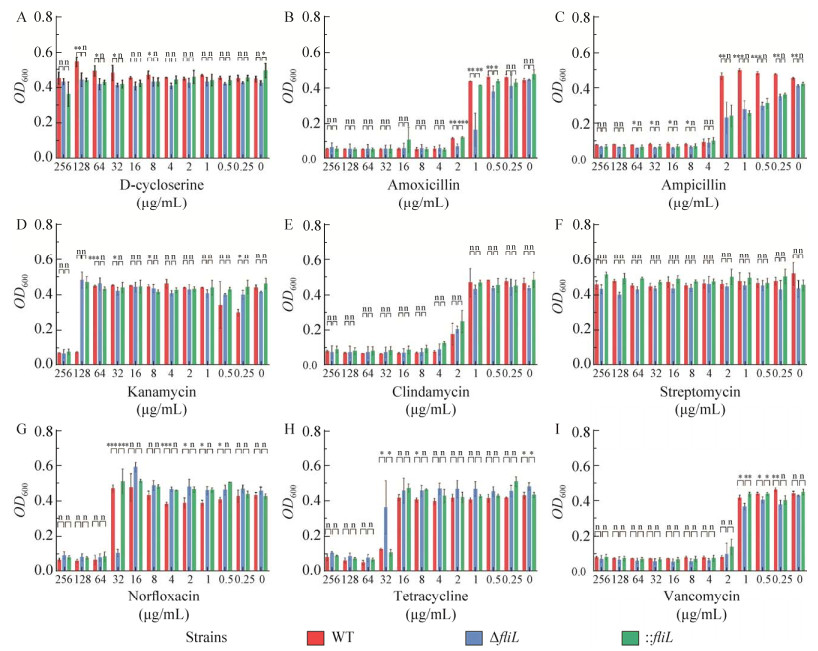
将菌株CD630、ΔfliL及: : fliL在含有不同种类及不同浓度的抗生素培养基中厌氧培养24 h后,测定OD600值后进行统计分析,与野生型CD630及回补菌株: : fliL相比,ΔfliL突变株对D环丝氨酸(D-cycloserine) (图 8A)、克林霉素(clindamycin) (图 8E)、链霉素(streptomycin) (图 8F)、万古霉素(vancomycin) (图 8I)等抗生素的敏感性无明显变化;ΔfliL对卡那霉素(kanamycin) (图 8D)和四环素(tetracycline) (图 8H)的敏感性降低;ΔfliL突变菌株对阿莫西林(amoxicillin) (图 8B)、氨苄青霉素(ampicillin) (图 8C)、诺氟沙星(norfloxacin) (图 8G)的敏感性提高。
|
| 图 8 野生型CD630、ΔfliL与: : fliL的抗生素敏感性测定 Fig. 8 Sensitivity of wild-type CD630, ΔfliL and : : fliL strains to antibiotics. A to I are antibiotics sensitivity profiles and statistical analyses of the CD630, ΔfliL, and : : fliL to different commonly used antibiotics. A: Sensitivity of strains to d-cycloserine. B: Sensitivity of strains to amoxicillin. C: Sensitivity of strains to ampicillin. D: Sensitivity of strains to kanamycin. E: Sensitivity of strains to clindamycin. F: Sensitivity of strains to streptomycin. G: Sensitivity of strains to norfloxacin. H: Sensitivity of strains to tetracycline. I: Sensitivity of strains to vancomycin. The ΔfliL mutant is more sensitive than the CD630 strain to amoxicillin, ampicillin, and norfloxacin, but it is more resistant than the CD630 to kanamycin and tetracycline. *: P < 0.05; **: P < 0.01; ***: P < 0.001; n: P > 0.05. |
| |
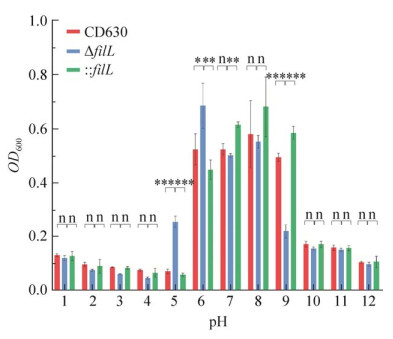
我们将野生型CD630菌株、ΔfliL及: : fliL分别接种于pH值为:1、2、3、4、5、6、7、8、9、10、11、12的BHIS培养基中,并在厌氧工作站中培养24 h后。测定OD600值后进行统计分析发现,在pH 1−4、pH 7−8和10−12之间,野生型CD630菌株、ΔfliL及: : fliL菌株pH耐受性无明显区别;在pH 5−6之间,ΔfliL菌株OD600值显著高于CD630菌株,: : fliL菌株OD600值回复至野生型水平;在pH 9时,ΔfliL菌株OD600值显著低于CD630菌株,: : fliL菌株OD600值回复至野生型水平。以上结果提示,ΔfliL菌株对弱酸环境耐受性增强;对弱碱性条件耐受性降低(图 9)。
|
| 图 9 野生型CD630、ΔfliL与: : fliL菌株pH耐受性 Fig. 9 pH tolerance of CD630, ΔfliL and : : fliL strains. The pH resistance and statistical analyses of strains CD630, ΔfliL and : : fliL. The red column represents the growth profile of CD630, the blue column represents the growth profile of fliL, the green column represents the growth profile of : : fliL. *: P < 0.05; **: P < 0.01; ***: P < 0.001; n: P > 0.05. |
| |
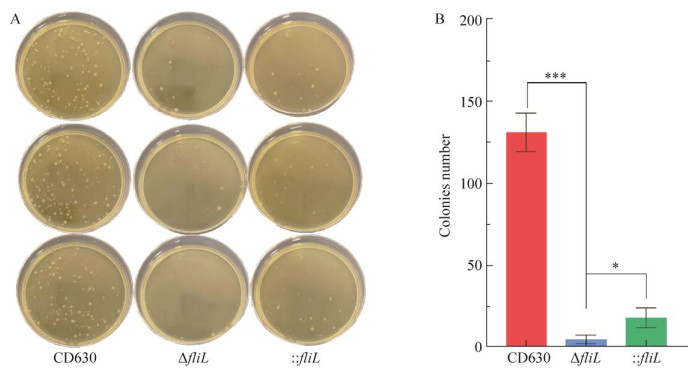
将野生型CD630菌株、ΔfliL及: : fliL置于60 ℃ 30 min条件下灭活营养细胞。灭活后的培养物收集后取20 μL分别接种于BHIS培养基并置于37 ℃厌氧培养5 d。孢子萌发后对平板上的菌落进行计数,ΔfliL菌株产生的孢子数量显著低于野生型CD630菌株(图 10),: : fliL菌株部分恢复了野生型表型。结果表明fliL基因在艰难拟梭菌产孢过程中具有重要的作用。
|
| 图 10 野生型CD630、ΔfliL及: : fliL产孢能力 Fig. 10 The sporulation abilities of wild-type CD630, ΔfliL and : : fliL strains. A: The colony forming units (CFU) of heat-shocked spore solution of strains CD630, ΔfliL and : : fliL on BHIS medium. B: The red column represents the CFU of CD630; the blue column represents the CFU of strain fliL; the green column represents the CFU of strain : : fliL. The sporulation ability of strains ΔfliL was significantly decreased than that of the WT and : : fliL strains. *: P < 0.05; ***: P < 0.001. |
| |
艰难拟梭菌为革兰阳性,产芽孢的机会致病性肠道微生物,是抗生素相关性腹泻和伪膜性肠炎的主要致病菌。艰难拟梭菌在人体肠道内进行运动及定殖等都在依赖鞭毛介导的游泳运动。有报道称,艰难拟梭菌的鞭毛突变体在仓鼠感染模型的体外和体内试验中显示出更高的毒性[23],由此可见鞭毛的功能可能对艰难拟梭菌致病性产生影响。FliL蛋白是影响鞭毛功能的单跨膜蛋白。弧菌属(Vibrio)中FliL蛋白与鞭毛基体相结合,影响鞭毛基体中的定子及转子的运动[6]。本研究中使用ACE技术对艰难拟梭菌的FliL蛋白编码基因fliL进行敲除,成功构建了突变菌株ΔfliL及回补菌株: : fliL。通过实验验证突变菌株ΔfliL与菌株CD630表型的差异,加深对艰难拟梭菌fliL基因功能的认识。我们测定了菌株CD630、ΔfliL及回补菌株: : fliL的生长曲线,结果证实fliL基因缺失后会影响艰难拟梭菌的最大生物量,除此之外ΔfliL突变株对于部分抗生素的敏感性也发生了变化。以上结果表明,艰难拟梭菌fliL基因可能在CDI的发生发展中发挥重要作用。
突变菌株ΔfliL对于不同作用机制的抗生素的耐受性出现不同程度的改变。其中,突变菌株ΔfliL对同为β-内酰胺类抗生素的阿莫西林及氨苄青霉素敏感性增高。Toth等研究发现艰难拟梭菌编码D类β内酰胺酶(CDD酶),其研究中cdd1基因在艰难拟梭菌自身启动子的作用下表达量不佳,相反将cdd1基因在匙形梭状杆菌(Clostridium cochlearium)的启动子作用下有效表达后,赋予了匙形梭状杆菌广谱的β-内酰胺类抗生素的耐受性[24]。研究表明临床上艰难拟梭菌对于喹诺酮类抗生素的耐受性主要由于喹诺酮耐药决定区(quinolone-resistance-determining region, QRDR)中内DNA旋转酶基因的突变从而导致艰难拟梭菌的蛋白发生变化[25],由此我们推测本研究中突变菌株ΔfliL对阿莫西林(图 8B)、氨苄青霉素(图 8C)及诺氟沙星(图 8G)敏感性提高,是由于fliL基因缺失影响了上述基因在艰难拟梭菌中的表达。另一方面,目前研究表明艰难拟梭菌中主要是tetM基因影响四环素的耐受性[26],fliL可能促进核糖体保护蛋白(TetM)的形成从而增强艰难拟梭菌对四环素的耐受性。卡那霉素所属的氨基糖苷类抗生素的耐受性改变可能的原因有:(1) 通过外排泵降低细菌细胞内抗生素的浓度;(2) 抗生素的分子靶标改变;(3) 氨基糖苷的酶失活[27]。由于ΔfliL对于卡那霉素敏感性降低,我们推测主要是由于fliL缺失后外排泵的作用增强而导致。
艰难拟梭菌的酸碱耐受性受到多种因素的影响,Wetzel等对环境pH值引起艰难拟梭菌生长繁殖、芽孢产生和萌发及艰难拟梭菌毒素量等生理变化进行了具体研究。人体肠道不同部位具有不同的酸碱度,艰难拟梭菌的定殖也与之息息相关。研究结果表明,环境pH值对于艰难拟梭菌毒素产生具有菌株依赖性差异,其中菌株630Δerm在pH值为6.5时产生的毒素最多,在pH值为5.5或pH为8.5时其生长及毒素产生显著减少[28]。本研究中,我们发现fliL基因缺失后ΔfliL菌株在pH为5−6之间OD600值显著高于CD630菌株,相反在pH为9时,ΔfliL菌株OD600值显著低于CD630菌株。fliL基因缺失后在偏酸及偏碱性的环境中都表现出了pH耐受性变化,提示fliL基因可影响艰难拟梭菌的酸碱耐受性,从而影响其在肠道环境中的竞争优势。
通过实验我们发现ΔfliL菌株相较于野生型,在0.3%半固体培养基上的迁移距离及迁移速度上均有显著下降。研究中将fliL基因回补至突变菌株后,可见回补菌株: : fliL在0.3%半固体培养基上的迁移距离及迁移速度,均有显著的上升。回补菌株: : fliL迁移距离甚至极显著地超过野生型,RT-qPCR发现fliL基因在艰难拟梭菌中表达量急剧升高。因此,我们推测: : fliL迁移距离极显著提高的原因与fliL基因在质粒上具有较高的拷贝数和表达剂量有关。艰难拟梭菌的鞭毛是游泳运动所必需的,鞭毛的运动可以帮助艰难拟梭菌向利于自身生存繁殖的环境进行迁移,除此之外有研究表明鞭毛有助于艰难拟梭菌在动物模型中粘附肠上皮细胞,并在定殖及其毒力上发挥作用[29-30]。鞭毛相关基因fliC及fliD突变株在仓鼠模型中的致病能力显著改变也验证了这一观点[30]。fliL基因位于鞭毛基体周围,具有维持艰难拟梭菌的运动能力。目前的研究证实fliL基因突变的变形杆菌运动能力大幅度降低,同时在小鼠模型中致病力大大降低[31]。本研究发现fliL基因的缺失极显著地降低了艰难拟梭菌的运动功能,由此可见fliL基因可能通过影响鞭毛基体转子及定子的运动影响其游泳能力。
细菌的运动性及芽孢都是影响细菌致病性的重要因素,两者之间可能存在一定的联系。Hashiguchi等[32]通过构建密苏里游动放线菌(Actinoplanes missouriensis)的FliA家族基因突变体验证了鞭毛相关的FliA家族基因在Actinoplanes missouriensis生成芽孢过程中的作用。结果显示ΔfliA1ΔfliA2突变体在孢子囊形成、孢子休眠方面存在缺陷,同时其产生的孢子量也较野生型有所下降,在ΔfliA2中甚至观察到游动孢子的游动速度降低,且突变菌株基因表达数据证实大量的鞭毛相关基因及转录调控因子表达量变化,其中fliL基因在ΔfliA2突变株中下调了5.2倍。以上结果推测在Actinoplanes missouriensis中fliL基因的敲除会影响其产孢基因的表达变化,从而影响其芽孢的生成过程。我们的研究结果证实了艰难拟梭菌fliL基因缺陷菌株ΔfliL在产孢能力上具有相似的缺陷,但其机制有待进一步的研究。
艰难拟梭菌的芽孢是细菌的休眠状态,艰难拟梭菌可以通过芽孢在恶劣的环境中保证自身的存活。在CDI期间,艰难拟梭菌启动孢子形成途径。休眠孢子产生后被人体摄入并在小肠中转变为代谢活跃状态,人体摄入艰难拟梭菌孢子后在小肠中转变为代谢活跃状态,在肠道代谢产物如胆汁酸的影响下转变为营养细胞。导致CDI在住院患者体内持续和传播[33]。一旦环境遭受其污染,很难通过消毒对其杀灭,孢子萌发后导致CDI复发[34]。除此之外,目前的研究表示芽孢形成与艰难拟梭菌的毒素产生是相关的,SigH及Spo0A因子已被证实参与了艰难拟梭菌生孢、孢子萌发过程[35-36]。我们在实验中发现菌株ΔfliL相较于CD630,其芽孢的产生及萌发出现明显地降低。上述结果提示FliL蛋白可能成为抑制艰难拟梭菌定殖及产孢能力的潜在靶点。
综上所述,fliL基因对于艰难拟梭菌的运动能力而言是必不可少的,并且显著影响了艰难拟梭菌芽孢的产生。除此之外flil基因还影响了艰难拟梭菌的生长,对于酸碱环境的适应能力。上述突变株的生理变化都可能在CDI发展及复发中导致严重的后果,因此fliL基因如何引起艰难拟梭菌表型的改变还需进一步在转录组水平、动物模型及分子程度上研究其相关信号通路的调控及引起组织的病理变化进行研究为后续研究临床CDI治疗药物提供理论基础。虽然本研究中对fliL基因缺失菌株的菌群进行了运动能力的探究,但是对于单个细菌鞭毛旋转的力量及其方向的研究尚存在空缺[37]。同时进一步研究fliL基因在鞭毛中的作用,从而揭示鞭毛蛋白对细菌运动能力的贡献,可为制备以鞭毛结构为基础的纳米机器等新兴领域提供有益参考。
| [1] |
吴媛, 李文革, 贾筱溪, 王媛媛, 张文竹. 我国艰难梭菌流行特征和研究进展. 疾病监测, 2021, 36(4): 319-323. WU Y, LI WG, JIA XX, WANG YY, ZHANG WZ. Epidemiological characteristics and research progress of Clostridioides difficile in China. Disease Surveillance, 2021, 36(4): 319-323 (in Chinese). |
| [2] |
HE M, MIYAJIMA F, ROBERTS P, ELLISON L, PICKARD DJ, MARTIN MJ, CONNOR TR, HARRIS SR, FAIRLEY D, BAMFORD KB, D'ARC S, BRAZIER J, BROWN D, COIA JE, DOUCE G, GERDING D, KIM HJ, KOH TH, KATO H, SENOH M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nature Genetics, 2013, 45(1): 109-113. DOI:10.1038/ng.2478
|
| [3] |
CZEPIEL J, DRÓŻDŻ M, PITUCH H, KUIJPER EJ, PERUCKI W, MIELIMONKA A, GOLDMAN S, WULTAŃSKA D, GARLICKI A, BIESIADA G. Clostridium difficile infection: review. European Journal of Clinical Microbiology & Infectious Diseases, 2019, 38(7): 1211-1221.
|
| [4] |
OFORI E, RAMAI D, DHAWAN M, MUSTAFA F, GASPERINO J, REDDY M. Community-acquired Clostridium difficile: epidemiology, ribotype, risk factors, hospital and intensive care unit outcomes, and current and emerging therapies. Journal of Hospital Infection, 2018, 99(4): 436-442. DOI:10.1016/j.jhin.2018.01.015
|
| [5] |
SANTIVERI M, ROA-EGUIARA A, KÜHNE C, WADHWA N, HU H, BERG HC, ERHARDT M, TAYLOR NMI. Structure and function of Stator units of the bacterial flagellar motor. Cell, 2020, 183(1): 244-257.e16. DOI:10.1016/j.cell.2020.08.016
|
| [6] |
TAKEKAWA N, ISUMI M, TERASHIMA H, ZHU SW, NISHINO Y, SAKUMA M, KOJIMA S, HOMMA M, IMADA K. Structure of Vibrio FliL, a new stomatin-like protein that assists the bacterial flagellar motor function. mBio, 2019, 10(2): e00292-e00219.
|
| [7] |
MOTALEB MA, PITZER JE, SULTAN SZ, LIU J. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. Journal of Bacteriology, 2011, 193(13): 3324-3331. DOI:10.1128/JB.00202-11
|
| [8] |
JENAL U, WHITE J, SHAPIRO L. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. Journal of Molecular Biology, 1994, 243(2): 227-244. DOI:10.1006/jmbi.1994.1650
|
| [9] |
SUASTE-OLMOS F, DOMENZAIN C, MIRELES-RODRÍGUEZ JC, POGGIO S, OSORIO A, DREYFUS G, CAMARENA L. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. Journal of Bacteriology, 2010, 192(23): 6230-6239. DOI:10.1128/JB.00655-10
|
| [10] |
ATTMANNSPACHER U, SCHARF BE, HARSHEY RM. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Molecular Microbiology, 2008, 68(2): 328-341. DOI:10.1111/j.1365-2958.2008.06170.x
|
| [11] |
CHAWLA R, FORD KM, LELE PP. Torque, but not FliL, regulates mechanosensitive flagellar motor-function. Scientific Reports, 2017, 7: 5565. DOI:10.1038/s41598-017-05521-8
|
| [12] |
MINTON NP, EHSAAN M, HUMPHREYS CM, LITTLE GT, BAKER J, HENSTRA AM, LIEW F, KELLY ML, SHENG L, SCHWARZ K, ZHANG Y. A roadmap for gene system development in Clostridium. Anaerobe, 2016, 41: 104-112. DOI:10.1016/j.anaerobe.2016.05.011
|
| [13] |
MCALLISTER KN, BOUILLAUT L, KAHN JN, SELF WT, SORG JA. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Scientific Reports, 2017, 7(1): 14672. DOI:10.1038/s41598-017-15236-5
|
| [14] |
ZHOU QS, RAO FQ, CHEN ZH, CHENG YM, ZHANG QF, ZHANG J, GUAN ZZ, HE Y, YU WF, CUI GZ, QI XL, HONG W. The cwp66 gene affects cell adhesion, stress tolerance, and antibiotic resistance in Clostridioides difficile. Microbiology Spectrum, 2022, 10(2): e0270421. DOI:10.1128/spectrum.02704-21
|
| [15] |
PURDY D, O'KEEFFE TAT, ELMORE M, HERBERT M, McLEOD A, BOKORI-BROWN M, OSTROWSKI A, MINTON NP. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Molecular Microbiology, 2002, 46(2): 439-452. DOI:10.1046/j.1365-2958.2002.03134.x
|
| [16] |
HEAP JT, EHSAAN M, COOKSLEY CM, NG YK, CARTMAN ST, WINZER K, MINTON NP. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Research, 2012, 40(8): e59. DOI:10.1093/nar/gkr1321
|
| [17] |
NG YK, EHSAAN M, PHILIP S, COLLERY MM, JANOIR C, COLLIGNON A, CARTMAN ST, MINTON NP. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One, 2013, 8(2): e56051. DOI:10.1371/journal.pone.0056051
|
| [18] |
YANG YY, SUN QQ, LIU Y, YIN HZ, YANG WP, WANG Y, LIU Y, LI YX, PANG S, LIU WX, ZHANG Q, YUAN F, QIU SW, LI J, WANG XF, FAN KQ, WANG WS, LI ZL, YIN SL. Development of a pyrF-based counterselectable system for targeted gene deletion in Streptomyces rimosus. Journal of Zhejiang University-SCIENCE B, 2021, 22(5): 383-396. DOI:10.1631/jzus.B2000606
|
| [19] |
EHSAAN M, KUEHNE SA, MINTON NP. Clostridium difficile genome editing using pyrE alleles[A]//Methods in Molecular Biology[M]. New York, NY: Springer New York, 2016: 35-52.
|
| [20] |
饶凤琴, 程玉梅, 吴昌学, 王义, 崔古贞, 齐晓岚, 洪伟. 敲除PaLoc毒力岛的无毒性艰难梭菌菌株的构建. 贵州医科大学学报, 2019, 44(10): 1128-1133. RAO FQ, CHENG YM, WU CX, WANG Y, CUI GZ, QI XL, HONG W. Knockout of PaLoc toxicity loci to construct non-toxic C. difficile strain. Journal of Guizhou Medical University, 2019, 44(10): 1128-1133 (in Chinese). |
| [21] |
LEE YY, BELAS R. Loss of FliL alters Proteus mirabilis surface sensing and temperature-dependent swarming. Journal of Bacteriology, 2015, 197(1): 159-173. DOI:10.1128/JB.02235-14
|
| [22] |
FLETCHER JR, PIKE CM, PARSONS RJ, RIVERA AJ, FOLEY MH, McLAREN MR, MONTGOMERY SA, THERIOT CM. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nature Communications, 2021, 12: 462. DOI:10.1038/s41467-020-20746-4
|
| [23] |
AUBRY A, HUSSACK G, CHEN WX, KUOLEE R, TWINE SM, FULTON KM, FOOTE S, CARRILLO CD, TANHA J, LOGAN SM. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infection and Immunity, 2012, 80(10): 3521-3532. DOI:10.1128/IAI.00224-12
|
| [24] |
TOTH M, STEWART NK, SMITH C, VAKULENKO SB. Intrinsic class D β-lactamases of Clostridium difficile. mBio, 2018, 9(6): e01803-e01818.
|
| [25] |
OH H, EDLUND C. Mechanism of quinolone resistance in anaerobic bacteria. Clinical Microbiology and Infection, 2003, 9(6): 512-517. DOI:10.1046/j.1469-0691.2003.00725.x
|
| [26] |
高琼, 黄海辉. 艰难梭菌耐药性及耐药机制研究进展. 遗传, 2015, 37(5): 458-464. GAO Q, HUANG HH. Update on antimicrobial resistance in Clostridium difficile. Hereditas, 2015, 37(5): 458-464 (in Chinese). DOI:10.16288/j.yczz.15-131 |
| [27] |
JANA S, DEB JK. Molecular understanding of aminoglycoside action and resistance. Applied Microbiology and Biotechnology, 2006, 70(2): 140-150. DOI:10.1007/s00253-005-0279-0
|
| [28] |
WETZEL D, McBRIDE SM. The impact of pH on Clostridioides difficile sporulation and physiology. Applied and Environmental Microbiology, 2020, 86(4): e02706-e02719.
|
| [29] |
BABAN ST, KUEHNE SA, BARKETI-KLAI A, CARTMAN ST, KELLY ML, HARDIE KR, KANSAU I, COLLIGNON A, MINTON NP. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One, 2013, 8(9): e73026. DOI:10.1371/journal.pone.0073026
|
| [30] |
TRZILOVA D, WARREN MAH, GADDA NC, WILLIAMS CL, TAMAYO R. Flagellum and toxin phase variation impacts intestinal colonization and disease development in a mouse model of Clostridioides difficile infection. Gut Microbes, 2022, 14(1): 2038854. DOI:10.1080/19490976.2022.2038854
|
| [31] |
李林俐, 朱来旭, 张雪, 费荣梅. 扬子鳄源普通变形杆菌FliL基因缺失株的构建及其特性分析. 南京农业大学学报, 2022, 45(4): 729-735. LI LL, ZHU LX, ZHANG X, FEI RM. Construction and characterization of Proteus vulgaris FliL gene deletion strain from Alligator sinensis. Journal of Nanjing Agricultural University, 2022, 45(4): 729-735 (in Chinese). |
| [32] |
HASHIGUCHI Y, TEZUKA T, OHNISHI Y. Involvement of three FliA-family sigma factors in the sporangium formation, spore dormancy and sporangium dehiscence in Actinoplanes missouriensis. Molecular Microbiology, 2020, 113(6): 1170-1188. DOI:10.1111/mmi.14485
|
| [33] |
CHIU CW, TSAI PJ, LEE CC, KO WC, HUNG YP. Inhibition of spores to prevent the recurrence of Clostridioides difficile infection-a possibility or an improbability?. Journal of Microbiology, Immunology and Infection, 2021, 54(6): 1011-1017. DOI:10.1016/j.jmii.2021.06.002
|
| [34] |
ALI S, MOORE G, WILSON APR. Spread and persistence of Clostridium difficile spores during and after cleaning with sporicidal disinfectants. The Journal of Hospital Infection, 2011, 79(1): 97-98. DOI:10.1016/j.jhin.2011.06.010
|
| [35] |
PETTIT LJ, BROWNE HP, YU L, SMITS WK, FAGAN RP, BARQUIST L, MARTIN MJ, GOULDING D, DUNCAN SH, FLINT HJ, DOUGAN G, CHOUDHARY JS, LAWLEY TD. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics, 2014, 15: 160. DOI:10.1186/1471-2164-15-160
|
| [36] |
SAUJET L, MONOT M, DUPUY B, SOUTOURINA O, MARTIN-VERSTRAETE I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. Journal of Bacteriology, 2011, 193(13): 3186-3196.
|
| [37] |
PARTRIDGE JD, NIETO V, HARSHEY RM. A new player at the flagellar motor: FliL controls both motor output and bias. mBio, 2015, 6(2): e02367.
|
 2023, Vol. 39
2023, Vol. 39