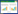中国科学院微生物研究所、中国微生物学会主办
文章信息
- 赵之怡, 张国强, 刘琨, 李盛英
- ZHAO Zhiyi, ZHANG Guoqiang, LIU Kun, LI Shengying
- 聚对苯二甲酸乙二醇酯水解酶研究进展
- Advances in poly(ethylene terephthalate) hydrolases
- 生物工程学报, 2023, 39(5): 1998-2014
- Chinese Journal of Biotechnology, 2023, 39(5): 1998-2014
- 10.13345/j.cjb.220915
-
文章历史
- Received: November 16, 2022
- Accepted: January 26, 2023
- Published: February 1, 2023
2. 青岛海洋科学与技术试点国家实验室 海洋生物学与生物技术功能实验室, 山东 青岛 266237
2. Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, Shandong, China
塑料因方便、耐用、成本低、可塑性高等优点被广泛应用于全球各地的生产、生活领域,极大地便捷了人类生活。但与此同时,塑料化学惰性高且极难降解,其大量使用造成了废弃物的严重积累[1]。目前全球每年生产约3.67亿t的塑料[2],其废弃物广泛存在于海洋、土壤、空气乃至人体中[3-5];经过海洋生物摄入[6],塑料在食物链中逐渐积累[7-8];大量塑料垃圾对生态环境、人类健康造成了巨大威胁[9-10]。聚对苯二甲酸乙二醇酯[poly(ethylene terephthalate), PET]是一类以对苯二甲酸(terephthalic acid, TPA)和乙二醇(ethylene glycol, EG)为单体,高度聚合的聚酯类高分子材料[8],在纺织产业、包装、食品工业等多个领域广泛应用[11-12]。据估计,世界上每分钟即有100万个PET瓶被生产出来[13]。对PET废弃物进行有效地回收处理对于减少资源浪费、维护生态稳定是迫在眉睫的。
传统的PET废弃物处理手段如填埋、焚烧、化学分解等方法成本较高,会产生多种有害分解产物,对环境产生二次污染[14-16];机械、热处理方法可用于PET塑料的回收再利用,但重复的回收处理会导致PET塑料性能下降[17]。因此,面对当前严峻的塑料污染,寻求新的PET废料的回收循环处理方法十分迫切。生物降解法具有反应环境温和、产生污染少、安全性高、能量消耗相对较低等突出优势,是一种极具前景的PET分解与再生策略[18]。近年来,基于PET水解酶的生物酶法降解展现出巨大的应用潜力[19],该方法通过PET水解酶催化PET的酯键水解,将长链分子分解为小分子物质,实现PET的降解与单体回收,从而实现PET的循环经济。本文旨在对该过程中最关键的PET水解酶的来源、催化活性、催化机理以及酶工程改造进行系统性综述,为PET的降解机制研究、PET高效降解酶的进一步挖掘和改造提供参考。
1 不同来源的PET水解酶 1.1 细菌来源的PET水解酶 1.1.1 来源于大阪伊德氏杆菌(Ideonella sakaiensis)的IsPETase及PET降解模型2016年,Yoshida等[20]在PET瓶回收厂中分离得到了一株能够降解PET的细菌(Ideonella sakaiensis) 201-F6,这是首次从自然界中分离获得能够以PET为唯一碳源生长的细菌,从该菌中发现了PET降解的关键酶——常温下具有最高PET水解活性的IsPETase,并首次揭示了PET的完整降解途径,这一针对传统意义上的“非可降解塑料”的生物降解研究引起了广泛的关注。该菌能够以低结晶度的PET膜为唯一碳源进行生长,根据该研究及相关文献[20-22]推测该菌降解PET的模型为“体外解聚,体内矿化”:(1) IsPETase通过N端信号肽介导的分泌途径被分泌到胞外,IsPETase表面疏水,附着于PET表面。长链PET分子进入IsPETase表面的催化裂隙被降解。PET长链被水解为对苯二甲酸双(2-羟乙基) [bis(2-hydroxyethyl) terephthalate, BHET]、对苯二甲酸单(2-羟乙基) [mono(2- hydroxyethyl) terephthalate, MHET]、TPA和EG等小分子产物(图 1A)。(2) 位于周质空间的MHETase将MHET分解为TPA和EG,进而通过中心代谢供给菌体生长所需(图 1B)。IsPETase在高温条件下稳定性较差,但温和条件下(30–40 ℃)对于PET的降解能力远高于TfH、LCC、FsC等其他对PET具有降解活性的酶,且对底物特异性较强,虽然许多酯类水解酶对碳链较短的底物更为偏好,但IsPETase对p-NP酰酯的活性低于TfH、LCC、FsC等,而对PET的降解活性却分别高出120、5.5、88倍[20]。
多种PET水解酶来源于嗜热放线菌,这部分酶的作用温度均在50−65 ℃之间,热稳定性较高。Kawai等[23]对(Saccharomonospora viridis) AHK190来源的Cut190进行了改造,其变体(S226P/R228S)的热稳定性得到提高,65 ℃条件下仍然非常稳定,70 ℃加热1 h后仍然存留40%的活性,在Ca2+存在时,63 ℃反应3 d,PET-GF膜的失重率达到13.5%±0.5%,PET-S膜失重率达到27.0%±1.0%。来源于(Thermobifida fusca) DSM43793的TfH (或称BTA1)在55 ℃的条件下降解经预处理的PET瓶,失重率达到近50%[24]。TfCut1、TfCut2 (T. fusca KW3来源)活性提高的突变体已经应用于涤纶纤维等纺织品表面的改性处理上[21]。除此之外,BTA2 (T. fusca DSM43793来源),Thc_Cut1、Thc_Cut2 (Thermobifida cellulosilytica DSM44535来源),Thf42_Cut1 (T. fusca DSM44342来源),Tha_Cut1 (T. fusca来源),Tcur_1278、Tcur_0390 (Thermomonospora curvata DSM43183来源),Thh_Est (Thermobifida halotolerans DSM44931来源)等也对不同PET底物表现出不同程度的降解能力(表 1)。
| PET hydrolases | Accession No. or PDB ID | Source | Reaction temperature/Substrate | PET degradation ability | References |
| BurPL | 7CWQ | Burkholderiales bacterium RIFCSPLOWO2_02_ FULL_57_36 | 30−40 ℃/mcPET, lcPET | Similar to IsPETase at 30 ℃, better than IsPETase at 40 ℃ | [28, 41-42] |
| BsEstB | HM040886 | Bacillus subtilis | 40 ℃/3PET | TPA, MHET and BA were generated | [43] |
| BTA2 | CAH17554.1 | Thermobifida fusca DSM43793 | 55−65 ℃/PET film | 48 h, 4% weight loss | [21, 44] |
| Cut190 | BAO42836.1 | Saccharomonospora viridis AHK190 | 50−65 ℃/PET-GFfilm, PET-S film | TPA can be detected | [23] |
| FsC | AAA33334.1 | Fusarium solani | 40 ℃/lcPET (crystallinity 7%) | 24 h, 40 ℃, 5%±1% weight loss | [29] |
| HiC | 4OYY | Humicola insolens | 70 ℃/lcPET (crystallinity 7%) | 96 h, 70 ℃, 97%±3% weight loss | [29] |
| IsPETase | GAP38373.1 | Ideonella sakaiensis 201-F6 | 20−45 ℃/PET film (crystallinity 1.9%) | 50 nmol/L, 30 ℃, 18 h, 0.3 mmol/L degradation product was generated | [20] |
| LCC | AEV21261.1 | Leaf-branch compost metagenome | 50−70 ℃/PET from plastic package | 50 ℃, degrades PET film at rate of 12 mg/h of enzyme | [33-34] |
| PES-H1/PHL7 | 7CUV/7NEI | Compost metagenome | 70 ℃/amorphous PET film | 16 h, 0.6 mgenzyme/gPET, weight loss > 90% | [35, 45] |
| PET2 | ACC95208.1 | Uncultured bacterium (marine metagenome) | 50 ℃/PET nanoparticle ager | Zone of clearance | [39] |
| PET5 | MBQ0729274.1 | Oleispira antarctica RB-8 | 50 ℃/PET nanoparticle ager | Zone of clearance | [39] |
| PET6 | UPI0003945E1F | Vibrio gazogenes | 50 ℃/PET nanoparticle ager | Zone of clearance | [39] |
| PET12 | A0A0G3BI90 | Polyangium brachysporum | 50 ℃/PET nanoparticle ager | Zone of clearance | [39] |
| PE-H | 6SBN | Pseudomonas aestusnigri VGXO14T (marine) | 30 ℃/amorphous PET film | 30 ℃, 48 h, (4.2±1.6) mg/L MHET was generated | [26] |
| Ple628 | 7VMD | Marine microbial consortium | 30 ℃/PET nanoparticles | 72 h, (52.9±1.1) µmol/L of MHET was released | [46] |
| Ple629 | 7VPA | Marine microbial consortium | 30 ℃/PET nanoparticles | 72 h, (785.9±27.8) µmol/L of MHET was released | [46] |
| RgPETase | 7DZT | Rhizobacter gummiphilus NS21 | 30−40 ℃/mc-PET and lcPET | Similar to IsPETase at 30 ℃ and 40 ℃ | [27] |
| SbPETase | AKJ29164.1 | Schlegelella brevitalea sp. nov. | 30 ℃/commercial PET film (crystallinity 10%) | The activity was one eighth that of IsPETase | [39, 47] |
| TfH/BTA1 | ALF04778.1 | Thermobifida fusca DSM43793 | 55 ℃/PET-B, PET-G | 55 ℃, 3 weeks, 50% weight loss of PET-B and PET-G | [24] |
| Thc_Cut2 | ADV92527.1 | Thermobifida cellulo- silytica DSM44535 | 50 ℃/PET film (crystallinity 37%) | 120 h, 5 mmol TA and 13 mmol MHET were generated per mole enzyme | [48-50] |
| Thc_Cut1 | ADV92526.1 | Thermobifida cellulo- silytica DSM44535 | 50 ℃/ PET film (crystallinity 37%) | 120 h, 56 mmol TA, 5 mmol MHET were generated per mole enzyme | [48-50] |
| Thf42_Cut1 | ADV92528.1 | Thermobifida fusca DSM44342 | 50 ℃ PET film (crystallinity 37%) | 120 h, 42 mmol TA and 6 mmol MHET were generated per mole enzyme | [48, 50] |
| Tha_Cut1 | ADV92525.1 | Thermobifida alba | 50 ℃/3PET | 50 ℃, 2 h, 0.6 μmol MHET, 2.6 μmol HEB and 0.3 μmol BA were generated | [24, 49-50] |
| Tcur_1278 | ACY96861.1 | Thermomonospora curvata DSM43183 | 50−55 ℃/PET nanoparticle | 80 μg/mL substrate, degrade rate 3.3×10–3 min–1 | [51-52] |
| Tcur_0390 | ACY95991.1 | Thermomonospora curvata DSM43183 | 50 ℃/PET nanoparticle | 20 μg/mL substrate, degrade rate 5.9×10–3 min–1 | [51-52] |
| Thh_Est | AFA45122.1 | Thermobifida halotolerans DSM44931 | 50 ℃/3PET | 19.8 mmol MHET and 1.5 mmol TPA were generated per mole enzyme | [53] |
| TfCut1 | CBY05529.1 | Thermobifida fusca KW3 | 55−65 ℃/lcPET film | 65 ℃, 48 h, Ca2+ exists, 11% weight loss | [21, 44] |
| TfCut2 | CBY05530.1 | Thermobifida fusca KW3 | 55−65 ℃/lcPET film | 65 ℃, 48 h, Ca2+ exist, 12.6% weight loss | [44] |
除备受关注的IsPETase以及来自于放线菌的PET水解酶外,许多来自其他细菌具有PET降解活性的酶相继被发现(表 1)。其中,来源于海洋微生物Pseudomonas aestusnigri VGXO14T中的PE-H[25]在30 ℃对无定型PET材料具有降解作用,将其250位酪氨酸替换为丝氨酸后,活性位点体积相比野生型扩大了一倍,其PET的降解活性得到了相应提高:底物为PETa (无定型PET材料)时,产生(5.4±0.6) mg/L MHET;底物为PETb (PET瓶材料)时,能检测到MHET的产生[26]。RgPETase分离自Rhizobacter gummiphilus,其与IsPETase对于微结晶度的PET (mc-PET)的降解活性相近,但对低结晶度的PET (lc-PET)降解能力较低[27]。BurPL分离自Burkholderiales bacterium,对PET的水解活性与IsPETase相近,但热稳定性更强[28],在40 ℃时的活性更高。除此之外,SbPETase、BsEstB等也来源于细菌,同样表现出对PET的降解活性(表 1)。
1.2 真菌来源的PET水解酶FsC (Fusarium solani来源)和HiC (Humicola insolens来源)两种PET水解酶来源于真菌,对PET具有不同程度的降解效果(表 1),主要降解产物为TPA与EG。FsC在50 ℃活性最强,与低结晶度PET膜(lcPET, 结晶度7%)孵育96 h得到5%的失重率。相比之下,HiC的热稳定性更高,且对底物具有更高的亲和性,在接近PET的玻璃化转变温度时(70 ℃)表现出最高活性,因而对PET的降解效率更高;虽然优先降解无定型PET膜,但其能力也足够降解lcPET,在70 ℃条件下,与底物孵育96 h可以实现97%±3%的失重率;对结晶度为37%的boPET也有一定降解能力[29]。
1.3 宏基因组来源的PET水解酶宏基因组策略于挖掘生态系统环境中不可培养部分的生物和基因多样性具有强大潜力,是从复杂的微生态系统中发现功能酶的有效手段[30-31]。设计合理高效的算法以探索有价值的宏基因组数据库、进行有效功能分析,对于发掘新基因具有重要意义[32]。LCC、PHL7 (PES- H1)、PET2、PET5、PET6、PET12、Ple628、Ple629等均来自环境宏基因组(表 1)。其中最具代表性的是LCC。
LCC分离自枝叶堆肥的宏基因组,是一种嗜热PET水解酶[33],受到了研究者的广泛关注。LCC结构中Cys275和Cys292之间形成二硫键,热稳定性强,最适反应温度为50 ℃;在65−70 ℃的温度下,24 h可降解24%−48%的低结晶性PET,溶解温度较高,85 ℃时仍然具有活力[34]。PES-H1 (PHL7)由Pfaff等分离自堆肥宏基因组,经过L92F/Q94Y的氨基酸替换后,PES-H1 (L92F/Q94Y)对无定型PET膜和生活中的商用PET的降解能力分别提高了2.3倍和3.4倍[35]。
一项有关于海洋宏基因组的研究表明,编码PET水解酶同源蛋白的基因分布于全球的海洋环境中[36],海洋微生物组对于塑料降解酶的开发具有巨大的潜力[37-38]。表 1所述PE-H、PET2、Ple628、Ple629均发现于海洋微生物。其中,PET2由Danso等[39]于海洋宏基因组中获得,其最适反应温度为55 ℃,在90 ℃孵育5 h后存留80%活性,热稳定性超过了LCC;Meyer-Cifuentes等[40]于海洋富集微生物宏基因组分离获得Ple628和Ple629,均能在30 ℃条件下降解PET纳米颗粒产生MHET,其中,Ple629的降解活性高于Ple628。
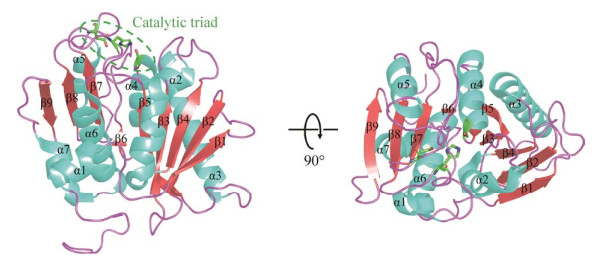
1.4 PET水解酶的进化关系与分类PET作为一种聚酯类塑料,其降解酶主要来自羧酸酯酶、脂肪酶和角质酶等水解酶[54-55],通过对表 1中的PET水解酶进行序列比对与系统发育树分析(图 2)可以看出,不同来源的PET水解酶在系统发育树中聚成了3个簇,即A簇、B簇和C簇。除了来自宏基因组且来源尚不明确的PSH-1和LCC外,A簇中的PET水解酶均来源于放线菌(革兰氏阳性菌);PSH-1和LCC与一系列革兰氏阳性菌来源的PET水解酶存在同一个簇中,进化关系较为接近,因而它们可能同样来源于革兰氏阳性菌。B簇中,IsPETase、RgPETase、BurPL、SbPETase均来自革兰氏阴性菌;PET2、PET5、PET6、PET12、Ple628和Ple629均来源于宏基因组,但除PET2、Ple628及Ple629的来源尚不明确,其余均被注释来自于革兰氏阴性菌;由于与一系列革兰氏阴性菌来源的PET水解酶进化关系接近,据此推测PET2、Ple628和Ple629可能同样来源于革兰氏阴性菌。C簇目前有3个PET水解酶,其中HiC、FsC均来自真菌,而BsEstB较为特殊,来自枯草芽孢杆菌,为革兰氏阳性菌,但其与真菌来源的HiC、FsC进化关系更为相近。通过系统发育树分析以及表 1中所示的降解活性来看,来自革兰氏阳性菌尤其是放线菌中的PET水解酶往往在高温下具有较高的活性。虽然在革兰氏阴性菌中发现的PET水解酶也会表现出一定的耐热性,如PET2、PET5、PET6、PET12可以在50 ℃表现出催化活性,但其降解能力却低于大部分革兰氏阳性菌来源的PET水解酶。以上分析可以为将来新型PET水解酶的发现提供一定的参考:耐热酶的发掘可重点关注革兰氏阳性细菌或真菌,而常温酶的发掘可重点关注革兰氏阴性细菌。
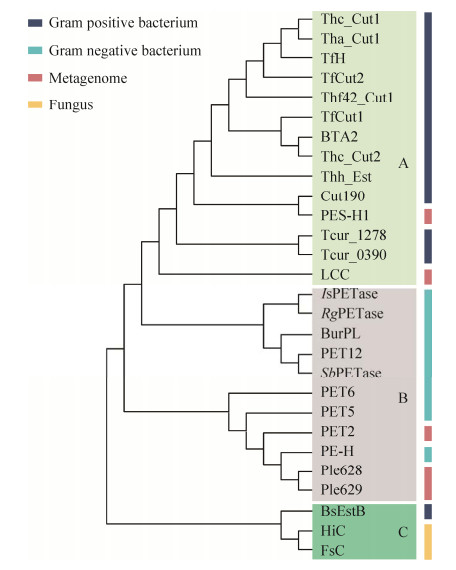
|
| 图 2 不同来源PET水解酶的系统发育树 Fig. 2 Phylogenetic tree of PET hydrolases from different sources generated by neighbor-joining algorithm. |
| |
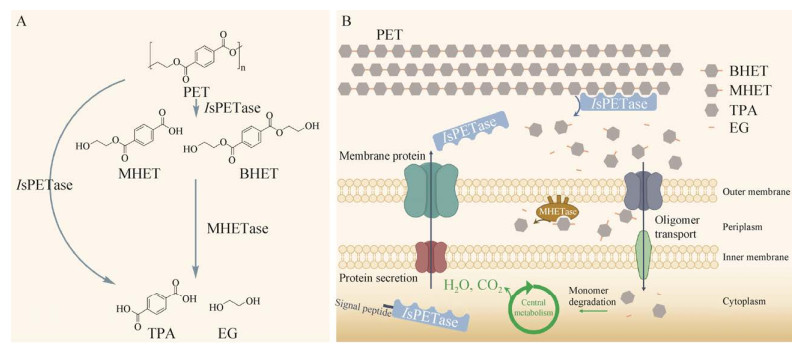
目前发现的大部分PET塑料降解酶同源性较为接近,大多属于α/β水解酶超家族,且其折叠结构也基本相同[56]。它们通常由两侧的α-螺旋和被其包裹着的几个相互平行的β-折叠构成,具有较保守的催化三联体残基序列[57-58],其对底物的降解过程基于催化三联体中作为亲核试剂的氨基酸残基对底物酯键的亲核攻击而进行[22]。
IsPETase的晶体结构于2017年被首次报道[59],其结构及对PET的降解机制受到广泛关注,相关研究在各类PET降解酶中最丰富[42, 60]。IsPETase同属于α/β水解酶超家族[61],中心结构中的9个β-折叠被7个α-螺旋所包围[62],具有保守的S160-H237-D206的催化三联体结构和氧阴离子穴,活性位点附近具有丝氨酸水解酶Gly– x1–Ser–x2–Gly基序(Gly158–Trp159–Ser160– Met161–Gly162)[39, 60](图 3)。IsPETase与TfH、LCC、Cut190等许多具有PET水解活性的酶高度同源,但同时也具有几个独特的结构特点。首先,IsPETase分子内有2个二硫键,其中特异的DS1连接着两个包括催化中心酸(D206)、碱(H237)的环。第二,其他同源酶中214位氨基酸为His,其侧链较大,且与W185距离更近,因此W185仅有1种构象,而IsPETase中214位氨基酸为Ser,其侧链较小,因而催化中心附近的W185在晶体结构中呈现出3种不同的构象,其中IsPETase W185所特有的B构象更利于PET底物与反应活性位点的结合。
目前已有多项研究对IsPETase催化PET降解的机制进行了解析。Han等[59]以2-羟乙基甲基对苯二甲酸酯[1-(2-hydroxyethyl) 4-methyl terephthalate, HEMT]、对硝基苯酚(para- nitrophenol, pNP)模拟反应底物,与IsPETase进行共结晶,得到了配体结合模型。在其研究中,第一个TPA单元内,HMET的酯氧原子与H237的侧链形成氢键,苯环与B构象的W185形成面—面堆积,羰基氧原子与构成氧阴离子洞的氨基酸M161、Y87的残基之间形成氢键,其他残基提供疏水相互作用;第二个TPA单元内,T88提供氢键作用力,W159提供苯环间的面—面堆积,因而HEMT被稳定结合于IsPETase中。pNP与TPA的结构非常类似,作者以pNP模拟TPA在IsPETase中的状态。pNP结合于HEMT的第一个TPA单元所结合的位点处,但相比HEMT,IsPETase与配体间的相互作用更少,仅有W185、I208、M161的疏水相互作用。其与IsPETase之间的相互作用力更弱,因而产物的苯甲酸基团可能与W185相互作用后,会被旋转并从催化中心释放[63]。
Joo等[60]以2-HE(MHET)4 [2-hydroxyethyl- (monohydroxyethyl terephthalate)4]为配体,通过分子对接,模拟了较长的PET链在IsPETase结合位点中的存在状态。Ser160、His237、Asp206构成了IsPETase的活性催化中心,其中Ser160作为亲核试剂攻击2-HE(MHET)4中的羰基碳原子,进而切断酯键。在其研究中,IsPETase的底物结合位点可以分为两部分,亚位点Ⅰ、亚位点Ⅱ。其中亚位点Ⅰ结合1个TPA单元,亚位点Ⅱ具有a、b、c这3个结合位点,分别结合3个TPA单元(图 4)。第一个MHET单体分子的苯环结合于Tyr87和Trp185两个芳香残基之间的沟壑上,主要以π-π键之间的相互作用稳定底物结合,同时Met161、Ile208为亚结合位点Ⅰ提供疏水的底面和侧面。亚结合位点Ⅱ相较位点Ⅰ更长、更浅,主要靠疏水作用力稳定底物结合。
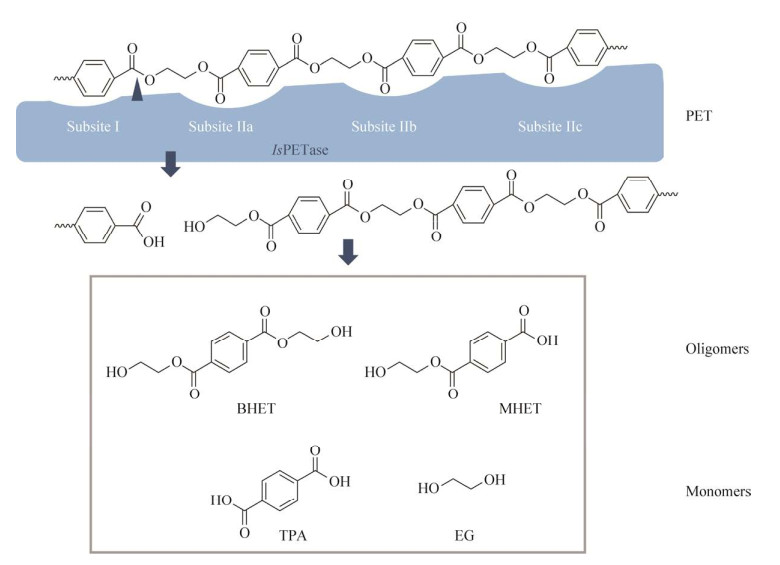
|
| 图 4 IsPETase催化PET水解机制推测 Fig. 4 Proposed mechanism for PET hydrolysis catalyzed by IsPETase. |
| |
IsPETase的催化活性中心位于亚位点Ⅰ和亚位点Ⅱ之间,催化第一个TPA单体与EG之间酯键的断裂,降解过程如图 4所示。IsPETase在胞外将PET分解为MHET等小分子物质,水解产物可能通过外膜蛋白(如孔蛋白)运输至周质空间,进而MHETase发挥作用,将MHET降解为PET单体物质[22](图 1B)。
3 PET水解酶的酶工程改造大多数PET水解酶的野生型仅对经过粉碎、结晶度低的PET底物具有较高的降解活性,且热稳定性较差,在中温环境中很快失去活性。因此,亟需针对PET水解酶的降解活性、热稳定性进行分子改造。许多研究基于蛋白结构分析,通过氨基酸替换,稳定活性位点、为底物的结合创造空间、提供疏水亲和力等实现活性位点的优化,或引入二硫键、分子内盐桥,稳定蛋白折叠结构,提高PET水解酶的性能[21]。
作为一种半结晶聚合物,PET含有结晶区和非晶态区。PET的玻璃化转变温度为75 ℃左右,高于75 ℃的环境将促进聚合物链的迁移,PET从结晶态转变为更活跃的无定形态[64],非晶态组分离开结晶区域,酶更容易接近聚合物链进行攻击,进而更有利于PET底物被水解[35, 55, 65]。PET酶解在水溶液中进行,在水环境中,链之间的相互作用减少、导致PET的Tg降低至60 ℃左右[66-67]。因此,60−75 ℃是较为适宜和高效的PET降解反应温度。大部分具有PET降解活性的酶在40−60 ℃范围内发挥作用,且即使部分水解酶可以在60−75 ℃的温度下工作,在高温条件下的长时间孵育同样容易丧失活性。因此有研究者针对稳定蛋白结构而进行分子改造,提高酶的热稳定性,使其在中温环境长时间保持高效降解能力。
3.1 IsPETase的酶工程改造IsPETase因其在常温下对PET相对较高的降解活性受到了较为广泛的研究。I. sakaiensis在温和环境中生存,因而IsPETase在常温下工作,虽然其对PET的降解活性普遍高于其他酶,但稳定性较差[68],在37 ℃孵育24 h后几乎失去活性[69],只能在远低于PET玻璃化转变温度的环境下工作;降解效果有限,离工业化应用仍有一定距离,50 nmol/L的IsPETase在30 ℃、pH 7.0的条件下与低结晶度PET膜反应18 h,只有约300 μmol/L的分解产物能够被检测到[20];其仅对低结晶度的PET材料具有相对高效的降解效果,对高结晶度的PET的降解能力极低。因此,有大量研究针对提高IsPETase的热稳定性、降解效率对其进行分子改造。
Son等[69]通过理性设计构建了IsPETase变体IsPETaseS121E/D186H/R280A,通过氨基酸替换,引入氢键作用力,极大地稳定了蛋白的折叠,使得IsPETaseS121E/D186H/R280A对PET的降解能力相比野生型提高了14倍,IsPETaseS121E/D186H/R280A也因此成为了许多研究者对IsPETase进行分子改造的起始酶[13, 70]。Cui等[71]通过基于结构的计算设计获得了DuraPETase,将Tm提高了31 ℃,相比野生型降解活性提高约300倍,对结晶度为30%的PET材料也表现出显著提高的降解能力。Bell等[13]通过定向进化筛选得到突变体HotPETase,其热稳定性极高,Tm高达82.5 ℃,在75 ℃孵育90 min仅有6%的酶活性损失。Lu等[54]通过基于结构的机器学习构建了IsPETase的突变体FAST-PETase,该突变体在50 ℃条件下表现出迄今为止最高的水解活性,相对IsPETaseS121E/D186H/R280A活性提高约38倍,可将商用消费后塑料(结晶度1.2%−6.2%)在1周内完全降解,作者通过FAST-PETase将着色后的商用PET材料解聚,回收降解产物TPA,进而重新聚合成为PET材料,实现了PET的生产-使用-回收-酶解-再生产的闭环。
通过结构分析、同源比对、定向进化等方法,S139T、W159H/S238F、W159H/F229Y、TS-PETase、TM3、D1等IsPETase突变体的水解活性、热稳定性也得到了不同程度提高,具体特性如表 2所示。
| Mutation sites | ΔTm (℃) | Degradation characteristics of PET (improved) | Design approach and interpretation |
References |
| S139T | NA* | Production amount of TPA increased by 8% | Directed evolution | [47] |
| W159H/S238F | +9.7 | Ability to reduce the crystallinity and products release amount was improved |
Homology modeling | [72] |
| S121E/D186H/R280A | +8.8 | 40 ℃, 72 h, PET degradation activity was increased by 14-fold |
Structure-based design | [69] |
| W159H/F229Y | +10.4 | 40 ℃, 24 h, PET degradation activity was increased by 40-fold |
Mutation design tool Premuse |
[73] |
| S214H/I168R/W159H/S188Q/ R280A/A180I/G165A/Q119Y/ L17F/T140D (DuraPETase) |
+31.0 | 37 ℃, 10 d, over 300-fold enhanced degradation activity toward PET films (crystallinity 30%) |
GRAPE strategy | [71] |
| S121E/D186H/R280A/N233C/ S282C (TS-PETase) |
+22.3 | Thermal stability was improved, and the activity retention time was prolonged at the reaction temperature. The products yield increased as the reaction proceeds |
Homology alignment | [74] |
| TS-PETase+K95N/F201I (TM3) | +5.3 | 120 times higher degradation of PET nanoparticles |
Directed evolution and structural comparison with LCC-ICCG mutant |
[70] |
| DuraPETase+N233C/S282C (D1) | +36.1 | Relative activity doubled at 50 ℃ and 60 ℃ | Structural comparison with LCC-ICCG mutant |
[70] |
| TS-PETase +P181V/S207R/S214Y/ Q119K/S213E/R90T/ Q182M/N212K/R224L/ S58A/S61V/K95N/M154G/ N241C/K252M/T270Q (HotPETase) |
+37.5 | Thermal stability was improved, generating 2.7×104 mol/L of product per mole of HotPETase within 1 h at 65 ℃, with degradability to commercial PET materials |
Directed evolution | [13] |
| S121E/D186H/R280A/R224Q/ N233K (FAST-PETase) |
+22.3 | Compared with S121E/D186H/R280A, PET hydrolysis activity increased by 2.4 and 38 times at 40 ℃ and 50 ℃ |
Structure-based, machine learning algorithm |
[54] |
| DuraPETase+N233K | +38.4 | IsPETase variant with the highest Tm so far | Structure-based, machine learning algorithm |
[54] |
| *: Not available. | ||||
LCC作为一种热稳定性强的水解酶,也是目前PET水解酶中的研究热点之一。Tournier等[75]在LCC的底物结合位点进行饱和突变,通过氨基酸替换引入二硫键(D238C/S283C),得到LCC-ICCG (F243I/D238C/S283C/Y127G),进一步提高了其热稳定性,Tm高达(70.1±1.5) ℃,可在72 ℃、10 h内将预处理后的PET瓶降解90%。2022年,Zeng等[76]进一步解析了LCC-ICCG的结构,并引入了3个位点突变A59K/V63I/N248P,可能通过介导β8-α6环的局部稳定,以及β-折叠中央和多个α-螺旋之间的稳定,使其Tm值进一步提高至98.9 ℃。
3.3 其他PET水解酶的酶工程改造其他PET水解酶通过基于结构的酶工程改造也获得了降解能力的提高。对于BurPL,研究者通过与IsPETase的结构比对分析引入了4个氨基酸替换(S335N/T338I/M363I/N365G),改造后BurPL的PET降解活性与热稳定性进一步提高,Tm值提高9 ℃,达到63 ℃,40 ℃条件下的PET水解活性提高2.8倍;野生型BurPL在40 ℃孵育24 h即失去一半酶活,经改造的酶在40 ℃孵育48 h仍然保持40%的活性[42]。对于TfCut2,Ren等[77]基于其和LCC的晶体结构,通过分子对接比较了两者底物结合袋的拓扑结构,在靠近PET二聚体模型底物的突变热点进行了工程改造,通过定点诱变产生TfCut2变体,对PET膜和纤维的水解活性显著增强,其中变体G62A和G62A/I213S,在65 ℃下反应50 h后,使PET膜失重近43%;Oda等[78]通过位点Asp250和Glu296引入一个二硫键,使酶的热稳定性得到极大提高,溶解温度提高了20−30 ℃以上。Pfaff等[35]对PES-H1进行了改造,L92F/Q94Y突变体对低结晶度PET粉末(结晶度13%)的降解能力高于野生型PES-H1及LCC-ICCG。
4 总结与展望目前,塑料制品的应用规模仍在不断扩大,且大部分塑料都为一次性使用产品,大量塑料废弃物在不断积累,渗透到自然环境、人类生活的方方面面。近年来,生物可降解塑料正在逐步发展[79],但就当下而言,因成本低、耐用、生产工艺成熟等突出优势,石油基塑料仍然是不可替代的。有研究预计,到2050年将有12亿t塑料垃圾堆积在垃圾填埋场或自然环境中[12],塑料污染成为亟待解决的问题。
以PET水解酶为代表的塑料生物降解近年来发展迅速并取得了突破性进展,从最开始仅能检测到微量降解产物,发展到实现完全解聚、回收单体并进行PET的再生产[54],或能实现通过菌株共培养进行PET废料发酵而发电、降解产物再合成PHB (聚羟基丁酸酯)等降解产物的再利用[80-81]。然而,对于产业化应用来说,PET的酶法降解仍然存在诸多限制:现有的降解酶对低结晶度PET降解效率较高,但对于高结晶度PET材料的降解能力极低[68];对于商用PET材料,需要经过充足的预处理(如粉碎、加热等)才能够实现有效分解[75],仍需消耗较多的能量;通过分子改造,降解酶突变体热稳定性的增加可能同时引起蛋白结构刚性的增加,热稳定性提升与水解活性的提高有时难以兼顾[70],二者之间应当存在较好的平衡。因此,挖掘新型高效、热稳定性高的PET水解酶对于高效回收PET废料具有重大意义。
虽然基于理性设计的PET水解酶改造近年来取得了很多成果[13, 54, 71, 75],但在突变体筛选过程中涉及大量的蛋白纯化以及产物的色谱分析,因此,提高酶活性检测效率、降低实验成本,对于高效PET水解酶的改造和开发十分关键。目前已有针对PET水解酶高通量筛选方法的研究报道,Wang等[47]基于提高酶的分泌表达量而建立了高通量筛选方法SecHTS,并筛选得到了活性提高的突变体IsPETaseS139T;Liu等[82]建立了基于双荧光检测的PET水解酶的高通量筛选方法,并通过筛选获得了6个活性提高的IsPETase突变体。因此,开发PET水解酶高通量筛选技术,高效评价大量降解酶的活性以实现快速筛选获得高效降解酶,将有助于实现更低成本的PET生物酶法降解回收。
| [1] |
LEBRETON L, ANDRADY A. Future scenarios of global plastic waste generation and disposal. Palgrave Communications, 2019, 5: 6. DOI:10.1057/s41599-018-0212-7
|
| [2] |
Plastics Europe Market Research Group (PEMRG) and Conversion Market & Strategy GmbH. Plastics Europe Plastics the Fact 2021 [EB/OL]. [2022-11-16]. https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/.
|
| [3] |
JENNER LC, ROTCHELL JM, BENNETT RT, COWEN M, TENTZERIS V, SADOFSKY LR. Detection of microplastics in human lung tissue using μFTIR spectroscopy. Science of the Total Environment, 2022, 831: 154907. DOI:10.1016/j.scitotenv.2022.154907
|
| [4] |
GREGORY MR. Environmental implications of plastic debris in marine settings—entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philosophical Transactions of the Royal Society B: Biological Sciences, 2009, 364(1526): 2013-2025. DOI:10.1098/rstb.2008.0265
|
| [5] |
EIAMTHONG B, MEESAWAT P, WONGSATIT T, JITDEE J, SANGSRI R, PATCHSUNG M, APHICHO K, SURARITDECHACHAI S, HUGUENIN-DEZOT N, TANG S, SUGINTA W, PAOSAWATYANYONG B, BABU MM, CHIN JW, PAKOTIPRAPHA D, BHANTHUMNAVIN W, UTTAMAPINANT C. Discovery and genetic code expansion of a polyethylene terephthalate (PET) hydrolase from the human saliva metagenome for the degradation and bio-functionalization of PET. Angewandte Chemie (International Ed in English), 2022, 61(37): e202203061.
|
| [6] |
CLERE IK, AHMMED F, REMOTO PIJG, FRASER- MILLER SJ, GORDON KC, KOMYAKOVA V, ALLAN BJM. Quantification and characterization of microplastics in commercial fish from southern New Zealand. Marine Pollution Bulletin, 2022, 184: 114121. DOI:10.1016/j.marpolbul.2022.114121
|
| [7] |
XU S, MA J, JI R, PAN K, MIAO AJ. Microplastics in aquatic environments: occurrence, accumulation, and biological effects. Science of the Total Environment, 2020, 703: 134699. DOI:10.1016/j.scitotenv.2019.134699
|
| [8] |
WEBB H, ARNOTT J, CRAWFORD R, IVANOVA E. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers, 2012, 5(1): 1-18. DOI:10.3390/polym5010001
|
| [9] |
YAN ZH, LIU YF, ZHANG T, ZHANG FM, REN HQ, ZHANG Y. Analysis of microplastics in human feces reveals a correlation between fecal microplastics and inflammatory bowel disease status. Environmental Science & Technology, 2022, 56(1): 414-421.
|
| [10] |
LIN SY, ZHANG HN, WANG C, SU XL, SONG YY, WU PF, YANG Z, WONG MH, CAI ZW, ZHENG CM. Metabolomics reveal nanoplastic-induced mitochondrial damage in human liver and lung cells. Environmental Science & Technology, 2022, 56(17): 12483-12493.
|
| [11] |
JAMBECK JR, GEYER R, WILCOX C, SIEGLER TR, PERRYMAN M, ANDRADY A, NARAYAN R, LAW KL. Marine pollution. Plastic waste inputs from land into the ocean. Science, 2015, 347(6223): 768-771. DOI:10.1126/science.1260352
|
| [12] |
GEYER R, JAMBECK JR, LAW KL. Production, use, and fate of all plastics ever made. Science Advances, 2017, 3(7): e1700782. DOI:10.1126/sciadv.1700782
|
| [13] |
BELL EL, SMITHSON R, KILBRIDE S, FOSTER J, HARDY FJ, RAMACHANDRAN S, TEDSTONE AA, HAIGH SJ, GARFORTH AA, DAY PJR, LEVY C, SHAVER MP, GREEN AP. Directed evolution of an efficient and thermostable PET depolymerase. Nature Catalysis, 2022, 5(8): 673-681. DOI:10.1038/s41929-022-00821-3
|
| [14] |
MARSHALL I, TODD A. The thermal degradation of polyethylene terephthalate. Transactions of the Faraday Society, 1953, 49: 67-78. DOI:10.1039/tf9534900067
|
| [15] |
PASZUN D, SPYCHAJ T. Chemical recycling of poly(ethylene terephthalate). Industrial & Engineering Chemistry Research, 1997, 36(4): 1373-1383.
|
| [16] |
GEYER B, LORENZ G, KANDELBAUER A. Recycling of poly(ethylene terephthalate)-a review focusing on chemical methods. Express Polymer Letters, 2016, 10(7): 559-586. DOI:10.3144/expresspolymlett.2016.53
|
| [17] |
RAHIMI A, GARCÍA JM. Chemical recycling of waste plastics for new materials production. Nature Reviews Chemistry, 2017, 1: 0046. DOI:10.1038/s41570-017-0046
|
| [18] |
SINGH JADAUN J, BANSAL S, SONTHALIA A, RAI AK, SINGH SP. Biodegradation of plastics for sustainable environment. Bioresource Technology, 2022, 347: 126697. DOI:10.1016/j.biortech.2022.126697
|
| [19] |
AHMED T, SHAHID M, AZEEM F, RASUL I, ALI SHAH A, NOMAN M, HAMEED A, MANZOOR N, MANZOOR I, MUHAMMAD S. Biodegradation of plastics: current scenario and future prospects for environmental safety. Environmental Science and Pollution Research, 2018, 25(8): 7287-7298. DOI:10.1007/s11356-018-1234-9
|
| [20] |
YOSHIDA S, HIRAGA K, TAKEHANA T, TANIGUCHI I, YAMAJI H, MAEDA Y, TOYOHARA K, MIYAMOTO K, KIMURA Y, ODA K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science, 2016, 351(6278): 1196-1199. DOI:10.1126/science.aad6359
|
| [21] |
CARR CM, CLARKE DJ, DOBSON ADW. Microbial polyethylene terephthalate hydrolases: current and future perspectives. Frontiers in Microbiology, 2020, 11: 571265. DOI:10.3389/fmicb.2020.571265
|
| [22] |
TANIGUCHI I, YOSHIDA S, HIRAGA K, MIYAMOTO K, KIMURA Y, ODA K. Biodegradation of PET: current status and application aspects. ACS Catalysis, 2019, 9(5): 4089-4105. DOI:10.1021/acscatal.8b05171
|
| [23] |
KAWAI F, ODA M, TAMASHIRO T, WAKU T, TANAKA N, YAMAMOTO M, MIZUSHIMA H, MIYAKAWA T, TANOKURA M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Applied Microbiology and Biotechnology, 2014, 98(24): 10053-10064. DOI:10.1007/s00253-014-5860-y
|
| [24] |
MÜLLER RJ, SCHRADER H, PROFE J, DRESLER K, DECKWER WD. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromolecular Rapid Communications, 2005, 26(17): 1400-1405. DOI:10.1002/marc.200500410
|
| [25] |
GOMILA M, MULET M, LALUCAT J, GARCÍA-VALDÉS E. Draft genome sequence of the marine bacterium Pseudomonas aestusnigri VGXO14T. Genome Announcements, 2017, 5(32): e00765-e00717.
|
| [26] |
BOLLINGER A, THIES S, KNIEPS-GRÜNHAGEN E, GERTZEN C, KOBUS S, HÖPPNER A, FERRER M, GOHLKE H, SMITS SHJ, JAEGER KE. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri-structural and functional insights. Frontiers in Microbiology, 2020, 11: 114. DOI:10.3389/fmicb.2020.00114
|
| [27] |
SAGONG HY, SON HF, SEO H, HONG H, LEE D, KIM KJ. Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis. Journal of Hazardous Materials, 2021, 416: 126075. DOI:10.1016/j.jhazmat.2021.126075
|
| [28] |
SAGONG HY, KIM S, LEE D, HONG H, LEE SH, SEO H, KIM KJ. Structural and functional characterization of an auxiliary domain-containing PET hydrolase from Burkholderiales bacterium. Journal of Hazardous Materials, 2022, 429: 128267. DOI:10.1016/j.jhazmat.2022.128267
|
| [29] |
RONKVIST ÅM, XIE WC, LU WH, GROSS RA. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules, 2009, 42(14): 5128-5138. DOI:10.1021/ma9005318
|
| [30] |
KIM DW, AHN JH, CHA CJ. Biodegradation of plastics: mining of plastic-degrading microorganisms and enzymes using metagenomics approaches. Journal of Microbiology, 2022, 60(10): 969-976. DOI:10.1007/s12275-022-2313-7
|
| [31] |
PUROHIT J, CHATTOPADHYAY A, TELI B. Metagenomic exploration of plastic degrading microbes for biotechnological application. Current Genomics, 2020, 21(4): 253-270.
|
| [32] |
DANSO D, CHOW J, STREIT WR. Plastics: environmental and biotechnological perspectives on microbial degradation. Applied and Environmental Microbiology, 2019, 85(19): e01095-e01019.
|
| [33] |
SULAIMAN S, YAMATO S, KANAYA E, KIM JJ, KOGA Y, TAKANO K, KANAYA S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Applied and Environmental Microbiology, 2012, 78(5): 1556-1562. DOI:10.1128/AEM.06725-11
|
| [34] |
SULAIMAN S, YOU DJ, KANAYA E, KOGA Y, KANAYA S. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry, 2014, 53(11): 1858-1869. DOI:10.1021/bi401561p
|
| [35] |
PFAFF L, GAO J, LI ZS, JÄCKERING A, WEBER G, MICAN J, CHEN YP, DONG WL, HAN X, FEILER CG, AO YF, BADENHORST CPS, BEDNAR D, PALM GJ, LAMMERS M, DAMBORSKY J, STRODEL B, LIU WD, BORNSCHEUER UT, WEI R. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catalysis, 2022, 12(15): 9790-9800. DOI:10.1021/acscatal.2c02275
|
| [36] |
ZRIMEC J, KOKINA M, JONASSON S, ZORRILLA F, ZELEZNIAK A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. mBio, 2021, 12(5): e0215521. DOI:10.1128/mBio.02155-21
|
| [37] |
GAO RR, SUN CM. A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. Journal of Hazardous Materials, 2021, 416: 125928. DOI:10.1016/j.jhazmat.2021.125928
|
| [38] |
GAO RR, LIU R, SUN CM. A marine fungus Alternaria alternata FB1 efficiently degrades polyethylene. Journal of Hazardous Materials, 2022, 431: 128617. DOI:10.1016/j.jhazmat.2022.128617
|
| [39] |
DANSO D, SCHMEISSER C, CHOW J, ZIMMERMANN W, WEI R, LEGGEWIE C, LI XZ, HAZEN T, STREIT WR. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Applied and Environmental Microbiology, 2018, 84(8): e02773-e02717.
|
| [40] |
MEYER-CIFUENTES IE, WERNER J, JEHMLICH N, WILL SE, NEUMANN-SCHAAL M, ÖZTÜRK B. Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nature Communications, 2020, 11(1): 5790. DOI:10.1038/s41467-020-19583-2
|
| [41] |
ANANTHARAMAN K, BROWN CT, HUG LA, SHARON I, CASTELLE CJ, PROBST AJ, THOMAS BC, SINGH A, WILKINS MJ, KARAOZ U, BRODIE EL, WILLIAMS KH, HUBBARD SS, BANFIELD JF. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nature Communications, 2016, 7: 13219. DOI:10.1038/ncomms13219
|
| [42] |
CHEN CC, HAN X, LI X, JIANG PC, NIU D, MA LX, LIU WD, LI SY, QU YY, HU HB, MIN J, YANG Y, ZHANG LL, ZENG W, HUANG JW, DAI LH, GUO RT. General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nature Catalysis, 2021, 4(5): 425-430. DOI:10.1038/s41929-021-00616-y
|
| [43] |
RIBITSCH D, HEUMANN S, TROTSCHA E, HERRERO ACERO E, GREIMEL K, LEBER R, BIRNER-GRUENBERGER R, DELLER S, EITELJOERG I, REMLER P, WEBER T, SIEGERT P, MAURER KH, DONELLI I, FREDDI G, SCHWAB H, GUEBITZ GM. Hydrolysis of polyethyleneterephthalate by p-nitrobenzylesterase from Bacillus subtilis. Biotechnology Progress, 2011, 27(4): 951-960. DOI:10.1002/btpr.610
|
| [44] |
THEN J, WEI R, OESER T, BARTH M, BELISÁRIO-FERRARI MR, SCHMIDT J, ZIMMERMANN W. Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnology Journal, 2015, 10(4): 592-598. DOI:10.1002/biot.201400620
|
| [45] |
SONNENDECKER C, OESER J, RICHTER PK, HILLE P, ZHAO ZY, FISCHER C, LIPPOLD H, BLÁZQUEZ-SÁNCHEZ P, ENGELBERGER F, RAMÍREZ-SARMIENTO CA, OESER T, LIHANOVA Y, FRANK R, JAHNKE HG, BILLIG S, ABEL B, STRÄTER N, MATYSIK J, ZIMMERMANN W. Low carbon footprint recycling of post-consumer PET plastic with a metagenomic polyester hydrolase. ChemSusChem, 2022, 15(9): e202101062.
|
| [46] |
MEYER CIFUENTES IE, WU P, ZHAO YP, LIU WD, NEUMANN-SCHAAL M, PFAFF L, BARYS J, LI ZS, GAO J, HAN X, BORNSCHEUER UT, WEI R, ÖZTÜRK B. Molecular and biochemical differences of the tandem and cold-adapted PET hydrolases Ple628 and Ple629, isolated from a marine microbial consortium. Frontiers in Bioengineering and Biotechnology, 2022, 10: 930140. DOI:10.3389/fbioe.2022.930140
|
| [47] |
WANG XT, SONG CY, QI QS, ZHANG YM, LI RJ, HUO LJ. Biochemical characterization of a polyethylene terephthalate hydrolase and design of high-throughput screening for its directed evolution. Engineering Microbiology, 2022, 2(2): 100020. DOI:10.1016/j.engmic.2022.100020
|
| [48] |
HERRERO ACERO E, RIBITSCH D, STEINKELLNER G, GRUBER K, GREIMEL K, EITELJOERG I, TROTSCHA E, WEI R, ZIMMERMANN W, ZINN M, CAVACO-PAULO A, FREDDI G, SCHWAB H, GUEBITZ G. Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules, 2011, 44(12): 4632-4640. DOI:10.1021/ma200949p
|
| [49] |
RIBITSCH D, ACERO EH, GREIMEL K, EITELJOERG I, TROTSCHA E, FREDDI G, SCHWAB H, GUEBITZ GM. Characterization of a new cutinase from Thermobifida alba for PET-surface hydrolysis. Biocatalysis and Biotransformation, 2012, 30(1): 2-9. DOI:10.3109/10242422.2012.644435
|
| [50] |
KAN YY, HE LH, LUO YZ, BAO R. IsPETase is a novel biocatalyst for poly(ethylene terephthalate) (PET) hydrolysis. Chembiochem, 2021, 22(10): 1706-1716. DOI:10.1002/cbic.202000767
|
| [51] |
WEI R, OESER T, THEN J, KÜHN N, BARTH M, SCHMIDT J, ZIMMERMANN W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express, 2014, 4: 44. DOI:10.1186/s13568-014-0044-9
|
| [52] |
CHERTKOV O, SIKORSKI J, NOLAN M, LAPIDUS A, LUCAS S, GLAVINA del RIO T, TICE H, CHENG JF, GOODWIN L, PITLUCK S, LIOLIOS K, IVANOVA N, MAVROMATIS K, MIKHAILOVA N, OVCHINNIKOVA G, PATI A, CHEN A, PALANIAPPAN K, DJAO ODN, LAND M, et al. Complete genome sequence of Thermomonospora curvata type strain (B9T). Standards in Genomic Sciences, 2011, 4(1): 13-22. DOI:10.4056/sigs.1453580
|
| [53] |
RIBITSCH D, HERRERO ACERO E, GREIMEL K, DELLACHER A, ZITZENBACHER S, MAROLD A, DIAZ RODRIGUEZ R, STEINKELLNER G, GRUBER K, SCHWAB H, GUEBITZ GM. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers, 2012, 4(1): 617-629. DOI:10.3390/polym4010617
|
| [54] |
LU HY, DIAZ DJ, CZARNECKI NJ, ZHU CZ, KIM W, SHROFF R, ACOSTA DJ, ALEXANDER BR, COLE HO, ZHANG Y, LYND NA, ELLINGTON AD, ALPER HS. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature, 2022, 604(7907): 662-667. DOI:10.1038/s41586-022-04599-z
|
| [55] |
SHIRKE AN, WHITE C, ENGLAENDER JA, ZWARYCZ A, BUTTERFOSS GL, LINHARDT RJ, GROSS RA. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: mechanism and effect on PET hydrolysis. Biochemistry, 2018, 57(7): 1190-1200. DOI:10.1021/acs.biochem.7b01189
|
| [56] |
李志帅, 高健, 陈纯琪, 郭瑞庭, 刘卫东, 韩旭. 聚对苯二甲酸乙二醇酯(PET)塑料水解酶结构、功能及改造. 生物加工过程, 2022, 20(4): 374-384. LI ZS, GAO J, CHEN CQ, GUO RT, LIU WD, HAN X. Structure, function and application of hydrolases for polyethylene terephthalate (PET) degradation. Chinese Journal of Bioprocess Engineering, 2022, 20(4): 374-384 (in Chinese). DOI:10.3969/j.issn.1672-3678.2022.04.003 |
| [57] |
JAEGER KE, RANSAC S, DIJKSTRA BW, COLSON C, van HEUVEL M, MISSET O. Bacterial lipases. FEMS Microbiology Reviews, 1994, 15(1): 29-63. DOI:10.1111/j.1574-6976.1994.tb00121.x
|
| [58] |
CHEN S, SU LQ, CHEN J, WU J. Cutinase: characteristics, preparation, and application. Biotechnology Advances, 2013, 31(8): 1754-1767. DOI:10.1016/j.biotechadv.2013.09.005
|
| [59] |
HAN X, LIU WD, HUANG JW, MA JT, ZHENG YY, KO TP, XU LM, CHENG YS, CHEN CC, GUO RT. Structural insight into catalytic mechanism of PET hydrolase. Nature Communications, 2017, 8: 2106. DOI:10.1038/s41467-017-02255-z
|
| [60] |
JOO S, CHO IJ, SEO H, SON HF, SAGONG HY, SHIN TJ, CHOI SY, LEE SY, KIM KJ. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nature Communications, 2018, 9: 382. DOI:10.1038/s41467-018-02881-1
|
| [61] |
RAUWERDINK A, KAZLAUSKAS RJ. How the same core catalytic machinery catalyzes 17 different reactions: the serine-histidine-aspartate catalytic triad of α/β-hydrolase fold enzymes. ACS Catalysis, 2015, 5(10): 6153-6176. DOI:10.1021/acscatal.5b01539
|
| [62] |
KAWAI F, KAWABATA T, ODA M. Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustainable Chemistry & Engineering, 2020, 8(24): 8894-8908.
|
| [63] |
CHEN CC, HAN X, KO TP, LIU WD, GUO RT. Structural studies reveal the molecular mechanism of PETase. The FEBS Journal, 2018, 285(20): 3717-3723. DOI:10.1111/febs.14612
|
| [64] |
ALVES NM, MANO JF, BALAGUER E, MESEGUER DUEÑAS JM, GÓMEZ RIBELLES JL. Glass transition and structural relaxation in semi-crystalline poly(ethylene terephthalate): a DSC study. Polymer, 2002, 43(15): 4111-4122. DOI:10.1016/S0032-3861(02)00236-7
|
| [65] |
MARTEN E, MÜLLER RJ, DECKWER WD. Studies on the enzymatic hydrolysis of polyesters. Ⅱ. Aliphatic-aromatic copolyesters. Polymer Degradation and Stability, 2005, 88(3): 371-381. DOI:10.1016/j.polymdegradstab.2004.12.001
|
| [66] |
BIANCHI R, CHIAVACCI P, VOSA R, GUERRA G. Effect of moisture on the crystallization behavior of PET from the quenched amorphous phase. Journal of Applied Polymer Science, 1991, 43(6): 1087-1089. DOI:10.1002/app.1991.070430608
|
| [67] |
LANGEVIN D, GRENET J, SAITER JM. Moisture sorption in pet influence on the thermokinetic parameters. European Polymer Journal, 1994, 30(3): 339-345. DOI:10.1016/0014-3057(94)90297-6
|
| [68] |
WALLACE NE, ADAMS MC, CHAFIN AC, JONES DD, TSUI CL, GRUBER TD. The highly crystalline PET found in plastic water bottles does not support the growth of the PETase-producing bacterium Ideonella sakaiensis. Environmental Microbiology Reports, 2020, 12(5): 578-582. DOI:10.1111/1758-2229.12878
|
| [69] |
SON HF, CHO IJ, JOO S, SEO H, SAGONG HY, CHOI SY, LEE SY, KIM KJ. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catalysis, 2019, 9(4): 3519-3526. DOI:10.1021/acscatal.9b00568
|
| [70] |
BROTT S, PFAFF L, SCHURICHT J, SCHWARZ JN, BÖTTCHER D, BADENHORST CPS, WEI R, BORNSCHEUER UT. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Engineering in Life Sciences, 2021, 22(3-4): 192-203.
|
| [71] |
CUI YL, CHEN YC, LIU XY, DONG SJ, TIAN YE, QIAO YX, MITRA R, HAN J, LI CL, HAN X, LIU WD, CHEN Q, WEI WQ, WANG X, DU WB, TANG SY, XIANG H, LIU HY, LIANG Y, HOUK KN, et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catalysis, 2021, 11(3): 1340-1350. DOI:10.1021/acscatal.0c05126
|
| [72] |
AUSTIN HP, ALLEN MD, DONOHOE BS, RORRER NA, KEARNS FL, SILVEIRA RL, POLLARD BC, DOMINICK G, DUMAN R, EL OMARI K, MYKHAYLYK V, WAGNER A, MICHENER WE, AMORE A, SKAF MS, CROWLEY MF, THORNE AW, JOHNSON CW, WOODCOCK HL, MCGEEHAN JE, et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(19): E4350-E4357.
|
| [73] |
MENG XX, YANG LX, LIU HQ, LI QB, XU GS, ZHANG Y, GUAN FF, ZHANG YH, ZHANG W, WU NF, TIAN J. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. International Journal of Biological Macromolecules, 2021, 180: 667-676. DOI:10.1016/j.ijbiomac.2021.03.058
|
| [74] |
ZHONG-JOHNSON EZL, VOIGT CA, SINSKEY AJ. An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Scientific Reports, 2021, 11: 928. DOI:10.1038/s41598-020-79031-5
|
| [75] |
TOURNIER V, TOPHAM CM, GILLES A, DAVID B, FOLGOAS C, MOYA-LECLAIR E, KAMIONKA E, DESROUSSEAUX ML, TEXIER H, GAVALDA S, COT M, GUÉMARD E, DALIBEY M, NOMME J, CIOCI G, BARBE S, CHATEAU M, ANDRÉ I, DUQUESNE S, MARTY A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature, 2020, 580(7802): 216-219. DOI:10.1038/s41586-020-2149-4
|
| [76] |
ZENG W, LI XQ, YANG YY, MIN J, HUANG JW, LIU WD, NIU D, YANG XC, HAN X, ZHANG LL, DAI LH, CHEN CC, GUO RT. Substrate-binding mode of a thermophilic PET hydrolase and engineering the enzyme to enhance the hydrolytic efficacy. ACS Catalysis, 2022, 12(5): 3033-3040. DOI:10.1021/acscatal.1c05800
|
| [77] |
REN W, OESER T, SCHMIDT J, RENÉ ME, BARTH M, THEN J, ZIMMERMANN W. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnology and Bioengineering, 2016, 113(8): 1658-1665. DOI:10.1002/bit.25941
|
| [78] |
ODA M, YAMAGAMI Y, INABA S, OIDA T, YAMAMOTO M, KITAJIMA S, KAWAI F. Enzymatic hydrolysis of PET: functional roles of three Ca2+ ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity. Applied Microbiology and Biotechnology, 2018, 102(23): 10067-10077. DOI:10.1007/s00253-018-9374-x
|
| [79] |
URBANEK AK, MIROŃCZUK AM, GARCÍA- MARTÍN A, SABORIDO A, DELA MATA I, ARROYO M. Biochemical properties and biotechnological applications of microbial enzymes involved in the degradation of polyester-type plastics. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2020, 1868(2): 140315.
|
| [80] |
LIU P, ZHANG T, ZHENG Y, LI QB, SU TY, QI QS. Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Engineering Microbiology, 2021, 1: 100003. DOI:10.1016/j.engmic.2021.100003
|
| [81] |
KALATHIL S, MILLER M, REISNER E. Microbial fermentation of polyethylene terephthalate (PET) plastic waste for the production of chemicals or electricity. Angewandte Chemie (International Ed in English), 2022, 61(45): e202211057.
|
| [82] |
LIU K, XU ZP, ZHAO ZY, CHEN YX, CHAI YT, MA L, LI SY. A dual fluorescence assay enables high-throughput screening for PET hydrolases. ChemSusChem, 2022, e202202019.
|
 2023, Vol. 39
2023, Vol. 39