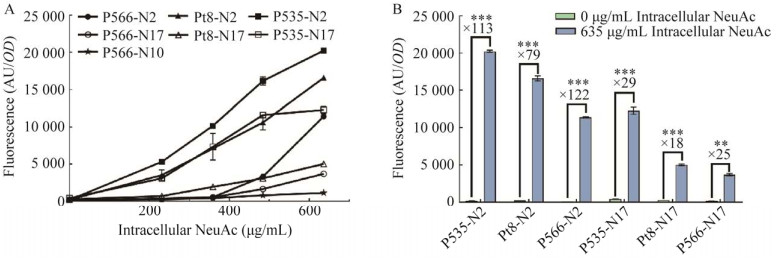
| [1] | |
|
| [2] |
RÖHRIG CH, CHOI SSH, BALDWIN N. The nutritional role of free sialic acid, a human milk monosaccharide, and its application as a functional food ingredient. Critical Reviews in Food Science and Nutrition, 2017, 57(5): 1017-1038. DOI:10.1080/10408398.2015.1040113
|
|
| [3] |
van KARNEBEEK CDM, BONAFÉ L, WEN XY, TARAILO-GRAOVAC M, BALZANO S, ROYER- BERTRAND B, ASHIKOV A, GARAVELLI L, MAMMI I, TUROLLA L, BREEN C, DONNAI D, CORMIER-DAIRE V, HERON D, NISHIMURA G, UCHIKAWA S, CAMPOS-XAVIER B, ROSSI A, HENNET T, BRAND-ARZAMENDI K, et al. NANS-mediated synthesis of sialic acid is required for brain and skeletal development. Nature Genetics, 2016, 48(7): 777-784. DOI:10.1038/ng.3578
|
|
| [4] |
WANG B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Advances in Nutrition, 2012, 3(3): 465S-472S. DOI:10.3945/an.112.001875
|
|
| [5] |
SALCEDO J, BARBERA R, MATENCIO E, ALEGRÍA A, LAGARDA MJ. Gangliosides and sialic acid effects upon newborn pathogenic bacteria adhesion: an in vitro study. Food Chemistry, 2013, 136(2): 726-734. DOI:10.1016/j.foodchem.2012.08.078
|
|
| [6] |
CHEESEMAN J, KUHNLE G, SPENCER DIR, OSBORN HMI. Assays for the identification and quantification of sialic acids: challenges, opportunities and future perspectives. Bioorganic & Medicinal Chemistry, 2021, 30: 115882.
|
|
| [7] |
ZHANG XL, LIU YF, LIU L, WANG M, LI JH, DU GC, CHEN J. Modular pathway engineering of key carbon-precursor supply-pathways for improved N-acetylneuraminic acid production in Bacillus subtilis. Biotechnology and Bioengineering, 2018, 115(9): 2217-2231. DOI:10.1002/bit.26743
|
|
| [8] |
LIU YF, LIU L, LI JH, DU GC, CHEN J. Synthetic biology toolbox and chassis development in Bacillus subtilis. Trends in Biotechnology, 2019, 37(5): 548-562. DOI:10.1016/j.tibtech.2018.10.005
|
|
| [9] |
YANG S, DU GC, CHEN J, KANG Z. Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis. Applied Microbiology and Biotechnology, 2017, 101(10): 4151-4161. DOI:10.1007/s00253-017-8142-7
|
|
| [10] |
NIU TF, LV XQ, LIU ZM, LI JH, DU GC, LIU L. Synergetic engineering of central carbon and nitrogen metabolism for the production of N-acetylglucosamine in Bacillus subtilis. Biotechnology and Applied Biochemistry, 2020, 67(1): 123-132. DOI:10.1002/bab.1845
|
|
| [11] |
YANG H, LIU YF, LI JH, LIU L, DU GC, CHEN J. Systems metabolic engineering of Bacillus subtilis for efficient biosynthesis of 5-methyltetrahydrofolate. Biotechnology and Bioengineering, 2020, 117(7): 2116-2130. DOI:10.1002/bit.27332
|
|
| [12] |
CARPENTER AC, PAULSEN IT, WILLIAMS TC. Blueprints for biosensors: design, limitations, and applications. Genes (Basel), 2018, 9(8): 375. DOI:10.3390/genes9080375
|
|
| [13] |
CAO YT, TIAN RZ, LV XQ, LI JH, LIU L, DU GC, CHEN J, LIU YF. Inducible population quality control of engineered Bacillus subtilis for improved N-acetylneuraminic acid biosynthesis. ACS Synthetic Biology, 2021, 10(9): 2197-2209. DOI:10.1021/acssynbio.1c00086
|
|
| [14] |
钱蕾, 刘延峰, 李江华, 刘龙, 堵国成. 适应性进化和改造质粒稳定性促进枯草芽孢杆菌合成 N-乙酰神经氨酸. 食品与发酵工业, 2021, 47(5): 1-6. QIAN L, LIU YF, LI JH, LIU L, DU GC. Regulating the synthesis of N-acetylneuraminic acid based on adaptive evolution and plasmid stability modification in Bacillus subtilis. Food and Fermentation Industries, 2021, 47(5): 1-6 (in Chinese).
|
|
| [15] |
DABIRIAN Y, GONÇALVES TEIXEIRA P, NIELSEN J, SIEWERS V, DAVID F. FadR-based biosensor- assisted screening for genes enhancing fatty acyl-CoA pools in Saccharomyces cerevisiae. ACS Synthetic Biology, 2019, 8(8): 1788-1800. DOI:10.1021/acssynbio.9b00118
|
|
| [16] |
LIU YF, ZHUANG YY, DING DQ, XU YR, SUN JB, ZHANG DW. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synthetic Biology, 2017, 6(5): 837-848. DOI:10.1021/acssynbio.6b00328
|
|
| [17] |
YEOM SJ, KIM M, KWON KK, FU YY, RHA E, PARK SH, LEE H, KIM H, LEE DH, KIM DM, LEE SG. A synthetic microbial biosensor for high- throughput screening of lactam biocatalysts. Nature Communications, 2018, 9: 5053. DOI:10.1038/s41467-018-07488-0
|
|
| [18] |
PANG QX, HAN H, LIU XQ, WANG ZG, LIANG QF, HOU J, QI QS, WANG Q. In vivo evolutionary engineering of riboswitch with high-threshold for N-acetylneuraminic acid production. Metabolic Engineering, 2020, 59: 36-43. DOI:10.1016/j.ymben.2020.01.002
|
|
| [19] |
PETERS G, de PAEPE B, de WANNEMAEKER L, DUCHI D, MAERTENS J, LAMMERTYN J, de MEY M. Development of N-acetylneuraminic acid responsive biosensors based on the transcriptional regulator NanR. Biotechnology and Bioengineering, 2018, 115(7): 1855-1865. DOI:10.1002/bit.26586
|
|
| [20] |
ZHANG XL, CAO YT, LIU YF, LIU L, LI JH, DU GC, CHEN J. Development and optimization of N-acetylneuraminic acid biosensors in Bacillus subtilis. Biotechnology and Applied Biochemistry, 2020, 67(4): 693-705. DOI:10.1002/bab.1942
|
|
| [21] |
EGAN M, O'CONNELL MOTHERWAY M, van SINDEREN D. A GntR-type transcriptional repressor controls sialic acid utilization in Bifidobacterium breve UCC2003. FEMS Microbiology Letters, 2015, 362(4): 1-9.
|
|
| [22] |
ZHANG XL, WANG CY, LV XQ, LIU L, LI JH, DU GC, WANG M, LIU YF. Engineering of synthetic multiplexed pathways for high-level N-acetylneuraminic acid bioproduction. Journal of Agricultural and Food Chemistry, 2021, 69(49): 14868-14877. DOI:10.1021/acs.jafc.1c06017
|
|
| [23] |
GIBSONDG, SMITH HO, HUTCHISON CA Ⅲ, VENTER JC, MERRYMAN C. Chemical synthesis of the mouse mitochondrial genome. Nature Methods, 2010, 7(11): 901-903. DOI:10.1038/nmeth.1515
|
|
| [24] |
ZHANG XZ, ZHANG YH P. Simple, fast and high-efficiency transformation system for directed evolution of cellulase in Bacillus subtilis. Microbial Biotechnology, 2011, 4(1): 98-105. DOI:10.1111/j.1751-7915.2010.00230.x
|
|
| [25] |
JEON Y, LEE YJ, KIM K, JANG G, YOON Y. Transcription factor-based biosensors for detecting pathogens. Biosensors, 2022, 12(7): 470. DOI:10.3390/bios12070470
|
|
| [26] |
UMENO D, KIMURA Y, KAWAI-NOMA S. Transcription factors as evolvable biosensors. Analytical Sciences, 2021, 37(5): 699-703. DOI:10.2116/analsci.20SCR12
|
|
| [27] |
LU ZH, YANG SH, YUAN X, SHI YY, OUYANG L, JIANG SJ, YI L, ZHANG GM. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis. Nucleic Acids Research, 2019, 47(7): e40. DOI:10.1093/nar/gkz072
|
|
| [28] |
LIU DY, MAO ZT, GUO JX, WEI LY, MA HW, TANG YJ, CHEN T, WANG ZW, ZHAO XM. Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synthetic Biology, 2018, 7(7): 1785-1797. DOI:10.1021/acssynbio.8b00115
|
|
| [29] |
YANG P, WANG J, PANG QX, ZHANG FY, WANG JS, WANG Q, QI QS. Pathway optimization and key enzyme evolution of N-acetylneuraminate biosynthesis using an in vivo aptazyme-based biosensor. Metabolic Engineering, 2017, 43: 21-28. DOI:10.1016/j.ymben.2017.08.001
|
|
| [30] |
RAMAN S, ROGERS JK, TAYLOR ND, CHURCH GM. Evolution-guided optimization of biosynthetic pathways. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(50): 17803-17808. DOI:10.1073/pnas.1409523111
|
|
 2023, Vol. 39
2023, Vol. 39