双加氧酶TET1表观遗传调控作用研究进展
徐凌
,
程钟坤
,
赵静贤
,
刘言言
,
赵永聚
,
杨晓伟
生物工程学报  2024, Vol. 40 2024, Vol. 40 Issue (12): 4351-4364 Issue (12): 4351-4364 |

DNA甲基化是一种重要的表观遗传修饰方式,在真核生物中DNA甲基化修饰主要发生在胞嘧啶第5位碳原子上,称为5-甲基胞嘧啶(5-methylcytosine, 5-mC),DNA甲基化在基因表达、基因沉默和基因组印记中起着重要作用[1-2]。然而,大多数的胞嘧啶甲基化是一个动态可逆的过程,TET1蛋白可以通过将5-甲基胞嘧啶迭代氧化成5-羟甲基胞嘧啶(5-hydroxymethylcytosine, 5-hmC)、5-甲酰基胞嘧啶(5-formylcytosine, 5-fC)和5-羧基胞嘧啶(5-carboxylcytosine, 5-caC),以及随后的碱基切除修复(base excision repair, BER)途径完成DNA的主动去甲基化过程[3]。TET1是最早发现的TET蛋白家族成员,其去甲基化作用对调控细胞基因转录表达具有重要意义,异常突变和表达失调的TET1蛋白与人类疾病,特别是与癌症的发病机制密切相关[4-5]。因此,本文综述了近年来双加氧酶TET1在各种生物学过程中发挥表观遗传调控作用的研究进展,分析了靶向TET1开发新的个性化药物治疗方案具有的潜在价值。
1 双加氧酶TET1概况 1.1 TET1蛋白去甲基化的发现与基本结构TET蛋白是生物体内普遍存在的一种α-KG和Fe2+依赖性双加氧酶。TET蛋白家族有3个成员,分别为TET1、TET2、TET3。最初TET蛋白是作为t(10;11)(q22;q23)异位急性髓系白血病(acute myeloid leukemia, AML)的融合蛋白被发现并命名,随后TET1又被发现能够在体外催化5-甲基胞嘧啶形成5-羟甲基胞嘧啶[6-7]。TET1基因位于染色体10q22,包含12个外显子,编码2 136个氨基酸[8]。TET1蛋白的N末端有1个Cys-Xaa-Xaa-Cys (CXXC)型锌指结构,所以该蛋白也被称为LCX (leukemia-associated protein with a CXXC domain)。而C末端的半胱氨酸富集区域(cysteine-rich domain, CD)、双链β-螺旋结构(double-stranded β-helix, DSBH)、Fe2+结合位点、α-KG结合位点共同组成了TET1的催化功能结构域。其中,CD和DSBH是TET1发挥催化活性所必需的。此外,其结构中还含有3个核定位信号(氨基酸: 20−50、620−653、1 158−1 162)能与DNA结合[9-12] (图 1)。值得注意的是,免疫代谢产物衣康酸能通过竞争的方式结合到TET2的α-KG结合位点,抑制TET2的催化功能[13]。由于TET蛋白家族具有相同的催化结构域[5],推测衣康酸或其他结构类似的代谢产物也可以通过与α-KG竞争结合位点而改变TET1生物学功能。
1.2 TET1蛋白去甲基化过程在哺乳动物基因组中,5-mC主要分布在胞嘧啶-磷酸-鸟嘌呤二核苷酸(CpG)中。TET1蛋白在α-KG和Fe2+的辅助下,将5-mC氧化成5-hmC。在此基础上,TET1还能将5-hmC氧化为5-fC,并再进一步氧化为5-caC。除了作为表观遗传标记,5-hmC还扮演了去甲基化中间体的角色。双加氧酶TET1的去甲基化的机制分为主动去甲基化和被动去甲基化这2类(图 2)。
主动去甲基化依赖于TET1对5-mC的氧化活性和BER途径。TET1首先将5-mC氧化为5-hmC并作为去甲基化中间产物:一方面,5-hmC在诱导活化脱氨酶(activation-induced deaminase, AID)和载脂蛋白B mRNA编辑催化蛋白(apolipoprotein B mRNA-editing enzyme catalysed polypeptide-like, APOBEC)的催化下生成5-羟甲基尿嘧啶(5-hydroxymethyluracil, 5-hmU),生成的5-hmU可被胸腺嘧啶DNA糖基化酶(thymine-DNA glycosylase, TDG)识别并切除,再通过BER重新转化为胞嘧啶C[14]。但这种机制仍存在争议,有研究表明,纯化出来的AID和APOBEC对5-mC的催化活性远远低于其常规底物胞嘧啶C,并且所有的AID和APOBEC家族成员都优先与未修饰的胞嘧啶C相互作用[15]。这一因素导致AID和APOBEC在DNA主动去甲基化过程中发挥的作用是有限的。另一方面,TET1将5-hmC催化为5-fC和5-caC,并再次通过TDG蛋白识别和切除基因组中的5-fC和5-caC,最后经过BER重新转化为胞嘧啶C,从而实现DNA的主动去甲基化[16]。值得注意的是,研究人员用15N标记5-caC后,发现5-caC可以在胚胎干细胞裂解产物的存在下直接脱羧并还原为未修饰的胞嘧啶C[17]。除此之外,细菌和哺乳动物的DNA甲基转移酶(DNA methyltransferases, DNMTs)也可以在体外催化5-caC直接脱羧生成未修饰的胞嘧啶C[18]。这些结果都为TET1催化生成5-caC进行后续的脱羧反应指明了新的研究方向。
DNA的被动去甲基化发生在DNA甲基化无法维持的情况下,随着DNA的复制,5-mC的含量逐渐被稀释。DNA甲基化主要依靠DNMTs来实现,其中,DNMT3a、DNMT3b和DNMT3c通过靶向未甲基化的CpG位点负责从头甲基化的建立,DNMT1在细胞分裂期间负责甲基化的维持[1]。在被动去甲基化过程中,TET1通过将DNA母链上的5-mC催化为5-hmC来阻碍DNMT1与新合成的子链结合,因为含有5-hmC的DNA双链作为底物与DNMT1结合会形成非生产性复合物,从而降低DNA甲基化的维持,最终通过DNA复制被动去除5-mC[19]。然而,这种机制的去甲基化效果并不显著,因为有研究人员将羟甲基化质粒和甲基化质粒分别稳定地转入到人体细胞中,发现二者在细胞复制和分裂过程中甲基化程度大致相等,这表明5-hmC并不能完全阻碍细胞中甲基化的维持[20]。
TET蛋白家族的DNA去甲基化作用主要发生在增强子、启动子及其他远端调节元件的调节区域[21]。在这些区域,TET蛋白家族可能与各种蛋白质和非编码RNA相互作用以发挥其调节功能。例如,GADD45A蛋白将TET1募集到CpG位点启动子上,通过其去甲基化作用介导肿瘤抑制因子TCF21的转录与激活[22]。除此之外,TET1的非催化活性也逐渐受到关注,例如,TET1和SIN3A之间相互作用以抑制内源性逆转录病毒元件,以及TET1通过srGAP3负向调控Neuro2a细胞的神经元分化并通过招募多聚酶抑制复合体2到染色质上以抑制原始内胚层标记Gata6、Sox17和滋养层外胚层标记Cdx2、Eomes的转录[23-25]。这些独立于其酶活性的非催化功能成为近年来研究的新方向。综上所述,TET1的催化和非催化活性特点均有助于其在不同细胞中调节基因的表达。
2 双加氧酶TET1表观遗传调控作用 2.1 调控胎盘及胎儿发育哺乳动物胚胎在发育过程中,会经历2次动态DNA去甲基化波动。第1次DNA去甲基化过程发生在受精后、胚胎植入前,母系和父系基因组在受精卵中发生了全局性的去甲基化过程[26]。第2次DNA去甲基化过程发生在胚胎植入后,原始生殖细胞(primordial germ cells, PGC)全基因组发生阶段和位点的特异性去甲基化,该过程的去甲基化状态主要由TET1/TET2介导的主动去甲基化调控完成[27-28]。在小鼠PGCs的发育中,TET1大量表达,其介导的5-mC氧化修饰对于正确擦除父系表达基因印记以及激活雌性生殖细胞中的减数分裂基因是必需的[29-31]。雄性小鼠的TET1基因被敲除后,其睾丸虽然具有正常形态和功能,但与正常雌性小鼠交配后,其主要的印记基因Peg10和Peg3等表达失调,引起后代产生胎盘发育不良、胎儿发育受限、出生后生长发育迟缓等表型;相应地,TET1敲除的雌性小鼠与正常雄性小鼠交配后会出现高达25%的胎儿致死率以及胎盘的发育异常[32]。而减数分裂基因Sycp1、Mael和Sycp3甲基化异常,引起卵母细胞减数分裂前期缺陷,造成卵母细胞大量凋亡,可能是导致TET1敲除雌性小鼠生育力下降的根本原因[33]。此外,TET1蛋白通过与关键的上皮型钙黏蛋白基因结合使其去甲基化来维持胎盘滋养层细胞上皮的完整性,敲除TET1的上皮组织更为松散,缺乏其原本的紧密特征[34]。另外,有一部分TET1/2基因联合缺失的细胞体积变大,出现多倍体滋养层巨细胞的形态特征,这可能是其在上皮间质转化过程中无法维持细胞周期蛋白B1稳定性而导致中心体复制和分离缺陷所造成的[33]。
TET1/2双基因敲除将导致严重的发育异常,包括外脑畸形和生长迟缓,并且引起围产期小鼠的死亡率上升[26]。TET1/3双基因敲除在E10.5之后无法检测到存活的胚胎,推测存在胚胎致死的现象[35]。TET1/2/3均敲除的小鼠胚胎致死率更高,其Nodal信号通路自E6.5之后表现出过分增强,这与其拮抗因子Lefty启动子高甲基化有关,最终导致小鼠中胚层和内胚层谱系分化失败而不能存活[36]。关于双加氧酶TET1在其中的作用,有研究发现在缺失TET1的上胚层细胞中,Lefty2显示出高甲基化且表达量减少,证实Lefty2是受TET1调控的基因[37]。为进一步研究TET1蛋白调控胚胎发育的作用,有研究人员将TET1蛋白的催化结构域和5′编码序列均切除,发现这些小鼠在原肠胚晚期表现出严重的前脑发育畸形和高死亡率,TET1功能缺陷小鼠细胞中Bin1、Cd44和Kirrel等驱动分化进程基因失调,导致更为明显的胚胎缺陷,证实了TET1在原肠胚形成前具有防止胚胎早熟分化的作用[37]。
2.2 调节免疫细胞活性当巨噬细胞受到脂多糖刺激时,细胞内的TET1将TNF-α启动子中5-mC转化为5-hmC以激活肿瘤坏死因子(tumour necrosis factor, TNF-α)基因的表达。利用基因编辑技术敲除巨噬细胞的TET1基因后,降低了TNF-α和其他炎性因子的表达[38]。除此之外,过敏性鼻炎患者的树突状细胞(dendritic cells, DCs)中双加氧酶TET1/TET3和5-hmC含量显著高于健康人员,过敏原刺激48 h后显著降低了DCs中TET1蛋白的表达,而敲除TET1后特应性DCs比非特应性DCs更容易被激活,细胞也更倾向于表达CD86、CD80和CD40[39]。同样地,在哮喘患者体内,TET1通过去甲基化作用直接调节干扰素和芳基烃受体信号通路的转录来抑制过敏性气道炎症[40]。以上这些结果表明,TET1可能在过敏性呼吸道炎症中发挥着重要作用。此外,在胸腺T细胞发育过程中,TET1/3通过增强子E4m和E4p的协同作用介导了Cd4位点的去甲基化及正常转录,这在CD4 T细胞阳性选择过程中是至关重要的[41]。并且,在Treg细胞分化过程中,TET1被转化生长因子-β (transforming growth factor-β, TGF-β)和白细胞介素-2 (interleukin-2, IL-2)募集到Foxp3位点,通过对其增强子CNS2的CpG位点去甲基化,稳定Foxp3表达,发挥调节Treg细胞的分化以及免疫稳态的作用[42-43]。另外,特异性降低小鼠子宫自然杀伤细胞中TET1表达,则会引起血管内皮生长因子C、干扰素-γ (interferon-γ, IFN-γ)和TGF-β等细胞因子表达水平发生显著性变化[44]。以上研究表明,TET1在调节免疫细胞活性方面发挥着不可或缺的作用。
2.3 重编程干细胞幼稚胚胎干细胞中富含5-hmC,但这种表观遗传修饰会随着其逐渐分化而迅速消失[45]。在多能胚胎干细胞中,参与产生5-hmC的主要蛋白TET1和TET2也会随着分化而迅速下调[45-46]。TET1/2双基因敲除的胚胎干细胞(embryonic stem cells, ESCs)会耗尽5-hmC并且更容易分化[34, 45, 47]。另外,TET1通过在与多能性密切相关基因Jarid2和Eed的mRNA上沉积5-hmC,降低mRNA的稳定性,调控多能性基因的适当表达,从而高效地维持细胞多能性分化过程[48]。此外,敲除TET1则会诱导DNMT3b的表达,降低端粒重组基因如Dmc1、Rad50和Smc1b的表达,从而表现出端粒缩短和染色体不稳定[49-50]。因此,TET1通过自身去甲基化功能作用于mRNA以及通过维持端粒稳定性来保证ESCs的自我更新能力和多能性潜力。TET1在小鼠ESCs的正常细胞周期进程和增殖中的还起着非催化作用,其主要机制为TET1抑制p21的表达进而调节DNA合成前期(G1期)到DNA合成期(S期)的快速进行[51]。
在骨髓间充质干细胞中,TET1的缺失会降低P2rX7启动子去甲基化水平,下调外泌体的释放,导致细胞内miR-297a-5p、miR-297b-5p和miR-297c-5p的聚集,影响骨髓间充质干细胞的自我更新及分化能力,出现明显的骨质疏松症表型[52]。脂肪组织中TET1介导DNA主动去甲基化激活并上调了Rxrα的表达,进而有利于间充质干细胞分化为脂肪细胞[53]。同时,TET1还能以非催化作用的方式抑制米色脂肪产热基因Ucp1、Ppargc1a的表达,进而影响细胞能量代谢状态[54]。在条件性敲除小鼠中,TET1的缺失导致海马齿状回颗粒下层中神经干细胞减少约45%,神经球表现出生长功能受损,参与神经干细胞增殖、神经保护、线粒体功能的基因Ng2、Ngb、Kctd14和Atp5h呈现高甲基化和下调表达状态[55]。在小鼠海马体神经干细胞中,TET1能直接与Dll3和Notch1启动子结合,催化启动子CpG序列中的5-mC羟基化为5-hmC,若上调TET1的表达则能激活海马体Notch信号传导通路,从而改善胎儿生长受限引起的神经干细胞减少及认知障碍[56]。
2.4 双向调控癌症发展DNA甲基化异常是肿瘤细胞的重要特征之一,并且TET蛋白家族介导的DNA去甲基化失衡是肿瘤致病的重要因素。5-mC和5-hmC与癌症的发生发展密切相关,而TET1蛋白在肿瘤发生中的作用机制也比较复杂,表现为双重调控作用。
TET1在淋巴血液系统恶性肿瘤中有着双重作用。如前文所述,TET1最初在AML中被鉴定为融合蛋白且在患者体内发挥致癌作用。在T细胞急性淋巴细胞白血病中TET1同样呈现出高度表达的状态,并通过维持5-hmC水平的催化特性调节RBPJ转录因子、Notch2和Notch3信号通路,以促进癌症的发展[57]。然而,敲除小鼠造血干细胞中的TET1后,则会在体外产生更多的具有自我更新能力的前体B细胞集落,在这些增殖的前体B细胞中显示出DNA断裂的异常增多并产生B细胞淋巴肿瘤,TET1的缺失促进了小鼠B细胞淋巴瘤的发展,揭示了TET1蛋白对B细胞淋巴瘤的潜在抑制作用[58]。
TET1在实体肿瘤方面同样具有双重作用。以乳腺癌为例,TET1被认为具有抑制乳腺癌肿瘤侵袭的作用。TET1蛋白对组织金属蛋白酶抑制因子2/3启动子的CpG序列去甲基化,增强其表达,下调金属蛋白酶的活性,进而抑制肿瘤细胞的侵袭[59]。但在三阴性乳腺癌中,TET1蛋白的表达显著增加,其在磷脂酰肌醇-3-羟激酶等致癌通路中导致CpG位点异常低甲基化从而激活通路发挥致癌作用[60-61]。此外,TET1与5-hmC还介导了一种过氧化氢依赖性的基因表达级联反应,驱动三阴性乳腺癌中肿瘤细胞的自我更新和增殖[62]。其次,TET1蛋白在肺癌中也发挥类似的作用。在非小细胞肺癌中,TET1蛋白和TDG结合在CD147启动子的同一区域,通过主动去甲基化上调其表达,从而促进肺癌的发展[63]。然而,在3-甲基胆蒽诱导的大鼠肺癌组织中TET1却显著下降,若特异性过表达TET1则在体内外均能显著抑制肺癌细胞的增殖、迁移和侵袭,TET1通过BER途径上调抑癌基因OGG1、XRCC1和APEX1启动子羟甲基化而实现抑制肺癌的作用[64]。
由此可见,在癌症的发生和发展过程中,TET1的调控机制是非常复杂的,对不同类型的肿瘤可表现为促进或抑制作用,或者在同一类型但不同亚型的肿瘤中发挥相反的作用。因此,靶向TET1蛋白的癌症治疗必须考虑这种双向作用,恢复致癌或抑癌基因启动子的正常甲基化水平可能成为预防和治疗某些癌症的替代策略。
2.5 保护神经细胞TET蛋白家族的表观遗传作用涉及成熟神经细胞类型的规范和包括神经元回路在内的高阶结构的发展,体现了其对大脑功能和神经发育至关重要的作用[65]。其中,TET1是神经系统中研究最多的TET家族成员。缺失TET1的小鼠表现出海马神经元受损,在莫里斯水迷宫实验中表现出空间学习和短期记忆能力的减退[55]。同样地,用siRNA敲除大鼠海马体中的TET1基因后,经Y迷宫训练形成的奖励记忆则受到了明显的抑制[66]。除此之外,TET1特异性缺失还会引起星形胶质细胞形态的异常并显著下调GluA1的表达进而损害Ca2+信号通路的转导,小鼠同样表现出学习和记忆能力的损害[67]。在缺血性脑卒中损伤中,TET1能通过氧调节蛋白150启动子去甲基化作用上调其表达,降低星形胶质细胞的凋亡和自噬,从而减轻脑卒中损伤[68]。另外,TET1还通过DNA去甲基化作用调节轴突-髓鞘界面离子转运的溶质载体蛋白SLC12A2的表达,这在小鼠少突胶质细胞的髓鞘修复过程中起着至关重要的作用[69-70]。这些研究表明,TET1参与大脑神经系统发育和疾病的发生发展,对维持神经系统的正常生理功能有着极其重要的作用。
值得注意的是,最近有研究发现TET1存在2个不同的异构体:TET1S (N末端截短转录本)和TETFL (经典全长转录本),二者在小鼠大脑从早期发育到成年的过程中出现差异性表达并发挥不同作用。TET1S在神经元中高度富集且与记忆形成相关,而TETFL在神经胶质细胞中含量更高并与神经细胞衰老、神经退行性疾病和癌症有关[71]。这表明不同异构体可能具有细胞类型特异性,并执行不同的功能,具体机制还有待深入研究。双加氧酶TET1在胚胎发育、免疫活性、干细胞调控、癌症发展和神经系统发育等过程中发挥的表观遗传调控作用详见表 1。
| Field | Epigenetic regulation | References |
| Embryogenesis | Demethylating the genome of the fertilized zygote Erasure of genomic imprinting Regulating meiotic gene expression Maintenance of trophoblast stem cells Critical for embryonic development |
[26-28] [30-32] [33] [34] [26, 35-37] |
| Immunocyte | Activator of TNF-α expression in macrophages Contribution to allergic airway inflammation Regulating differentiation of T cells Affecting proliferation of mouse uterine natural killer cells and the transcriptional level |
[38] [39-40] [41-43] [44] |
| Stem cell | Establishment of pluripotency and differentiation Regulating telomere maintenance and chromosomal stability Maintenance mesenchymal stem cells Promotor of RXRα expression and adipogenesis Regulating hippocampal neurogenesis and cognition |
[34, 45-48] [49-50] [52] [53] [55-56] |
| Cancer | Promotor of AML Suppressor of B cell lymphoma Dual function in breast and lung cancer |
[7, 57] [58] [59-64] |
| Neuron | Critical for brain function and cognition formation Regulating astrocyte development and adult remyelination |
[55, 66-67] [68-70] |
| Others (non-catalysis) |
Ensuring proper cell cycle progression Suppressor of beige adipocyte |
[51] [54] |
TET1功能的发挥也受到不同因素的影响(图 3)。TET1是α-KG和Fe2+依赖性的双加氧酶,细胞内α-KG和Fe2+的改变将直接影响其活性。外源饲喂1-硝基芘可以显著降低小鼠胚胎前脑中α-KG的含量,导致TET1的酶活性降低[72]。此外,用α-KG的结构相似产物2-羟基戊二酸(2-hydroxyglutaric acid, 2-HG)处理TH17细胞后,TET1的酶活性受到抑制,同时其转录因子Foxp3的甲基化水平上调,表达量下降[73]。与α-KG结构相似的琥珀酸和富马酸,同样可以对TET1产生抑制作用[13]。除此之外,维生素C已被证明能显著增加成纤维细胞转化成多能干细胞过程中5-hmC及其氧化衍生物5-fC、5-caC的产生,而这种功能需要依赖TET酶才能实现[8, 74-75]。维生素C通过与TET蛋白C末端催化结构域的直接相互作用,增强小鼠胚胎成纤维细胞5-hmC的产生,而其作用机制可能是促进了TET蛋白的辅助因子Fe2+的折叠或再循环[76-77]。另外,含有芳香族羟基的腙是一类金属螯合剂,能通过络合TET1催化结构位点上的Fe2+形成复合物,降低TET1的酶活性[78]。
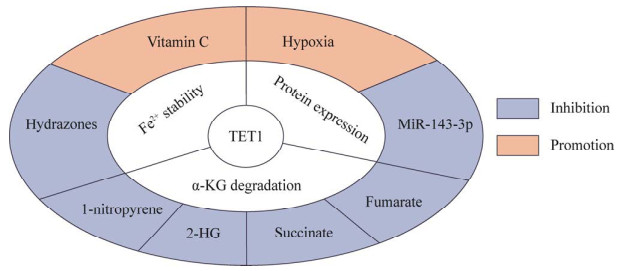
|
| 图 3 TET1的上游调控 Fig. 3 Upstream regulation of TET1. 2-HG: 2-hydroxyglutaric acid. |
| |
在患有大脑中动脉闭塞小鼠的星形胶质细胞中,miR-143-3p异常增加可以下调TET1的表达,促进星形胶质细胞凋亡和缺血性脑损伤,而靶向降低星形胶质细胞中miR-143-3p水平则会引起TET1的表达显著增高,这表明miR-143-3p可以负向调节细胞中TET1的表达[68]。此外,TET1蛋白的CXXC区域还通过与缺氧诱导因子HIF-1α和HIF-2α的相互作用,以非催化方式充当共激活因子并促进上皮-间充质转化[79]。2%的低氧环境下,TET1在脂肪细胞中表达水平显著提高,诱导瘦素和IL-6、IL-1β、IFN-γ等炎性细胞因子的分泌[80],提示细胞微环境如低氧也是调控TET1蛋白活性的重要因素。
4 总结与展望作为最早被发现的双加氧酶家族成员,TET1在哺乳动物胚胎发育、免疫细胞活性、干细胞重编、癌症调控以及空间学习等方面发挥的表观遗传功能已被大量研究证实,并且在过去的几年里,TET1蛋白功能的失调已显示出与多种疾病的发生有着不可忽视的因果关系。在不同的细胞背景、细胞代谢状态、细胞微环境及转录后修饰等影响因素下,TET1的作用方式不同,其结果也有所不同。因此,深入研究TET1的作用机制对癌症的防控、生殖发育的健康发展以及再生医学具有重要的意义。
虽然TET1是近些年表观遗传学研究重点,有关其作用机制的研究结果不断增加,但其所介导的去甲基化是一个非常复杂的表观遗传调控体系,仍存在许多需要继续探索的问题。如TET1的肿瘤促进和抑制作用之间是否存在作用平衡机制,这对利用其活性筛选新的癌症治疗靶点以及进行恶性肿瘤等疾病的早期预测至关重要。此外,TET1非催化活性及其与催化活性的潜在共同作用机制尚不清楚,先前的研究大多指向TET1在表观遗传调控中发挥的催化活性,但其非催化活性在一些生理及病理过程中的作用尚未阐明,许多实验模型仅排除了TET1催化结构域的作用,却很少将其非催化活性以及二者的潜在共同作用考虑在内,尤其是在胚胎发育方面。目前,已有部分研究尝试应用TET蛋白抑制剂等技术储备来治疗疾病,但是由于TET1在各种病理反应中调节机制的复杂性,这类治疗手段仍需更多体内外模型的验证,通过更准确的实验数据才能证明其治疗效果。另外,2-羟基戊二酸、琥珀酸、富马酸以及衣康酸等细胞代谢产物均具有调控TET1活性的作用,结合TET1对细胞代谢调控作用的研究结果,提示TET1与细胞代谢之间存在相互调控的关系,而二者互作关系的平衡机制仍待探究。因此,通过代谢学、肿瘤学以及表观遗传学等多学科交叉研究,全面深入地揭示双加氧酶TET1的生物活性及其调控机制,是其进行生物学应用的首要问题。
| [1] |
CHEN ZY, ZHANG Y. Role of mammalian DNA methyltransferases in development[J]. Annual Review of Biochemistry, 2020, 89: 135-158. DOI:10.1146/annurev-biochem-103019-102815
|
| [2] |
GREENBERG MVC, BOURC'HIS D. The diverse roles of DNA methylation in mammalian development and disease[J]. Nature Reviews Molecular Cell Biology, 2019, 20: 590-607. DOI:10.1038/s41580-019-0159-6
|
| [3] |
KRIUKIENĖ E, TOMKUVIENĖ M, KLIMAŠAUSKAS S. 5-hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA[J]. Chemical Society Reviews, 2024, 53(5): 2264-2283. DOI:10.1039/D3CS00858D
|
| [4] |
NISHIYAMA A, NAKANISHI M. Navigating the DNA methylation landscape of cancer[J]. Trends in Genetics, 2021, 37(11): 1012-1027. DOI:10.1016/j.tig.2021.05.002
|
| [5] |
MA CL, SEONG H, LIU YM, YU X, XU SL, LI YJ. Ten-eleven translocation proteins (TETs): tumor suppressors or tumor enhancers?[J]. Frontiers in Bioscience (Landmark Edition), 2021, 26(10): 895-915. DOI:10.52586/4996
|
| [6] |
RASMUSSEN KD, HELIN K. Role of TET enzymes in DNA methylation, development, and cancer[J]. Genes & Development, 2016, 30(7): 733-750.
|
| [7] |
TAHILIANI M, KOH KP, SHEN YH, PASTOR WA, BANDUKWALA H, BRUDNO Y, AGARWAL S, IYER LM, LIU DR, ARAVIND L, RAO A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1[J]. Science, 2009, 324(5929): 930-935. DOI:10.1126/science.1170116
|
| [8] |
MOHR F, DÖHNER K, BUSKE C, Rawat VPS. TET genes: new players in DNA demethylation and important determinants for stemness[J]. Experimental Hematology, 2011, 39(3): 272-281. DOI:10.1016/j.exphem.2010.12.004
|
| [9] |
KIM H, JANG WY, KANG MC, JEONG J, CHOI M, SUNG Y, PARK S, KWON W, JANG S, KIM MO, KIM SH, RYOO ZY. TET1 contributes to neurogenesis onset time during fetal brain development in mice[J]. Biochemical and Biophysical Research Communications, 2016, 471(4): 437-443. DOI:10.1016/j.bbrc.2016.02.060
|
| [10] |
HUANG SS, ZHU ZQ, WANG YQ, WANG YR, XU LX, CHEN XM, XU Q, ZHANG QM, ZHAO X, YU Y, WU DL. Tet1 is required for Rb phosphorylation during G1/S phase transition[J]. Biochemical and Biophysical Research Communications, 2013, 434(2): 241-244. DOI:10.1016/j.bbrc.2013.02.110
|
| [11] |
ZHANG WY, LU ZP, GAO YE, YE LH, SONG TQ, ZHANG XD. MiR-520b suppresses proliferation of hepatoma cells through targeting ten-eleven translocation 1 (TET1) mRNA[J]. Biochemical and Biophysical Research Communications, 2015, 460(3): 793-798. DOI:10.1016/j.bbrc.2015.03.108
|
| [12] |
YANG JH, BASHKENOVA N, ZANG RG, HUANG X, WANG JL. The roles of TET family proteins in development and stem cells[J]. Development, 2020, 147(2): dev183129. DOI:10.1242/dev.183129
|
| [13] |
CHEN LL, MORCELLE C, CHENG ZL, CHEN XF, XU YP, GAO YJ, SONG JB, LI ZJ, SMITH MD, SHI M, ZHU YZ, ZHOU N, CHENG M, HE CX, LIU K, LU GP, ZHANG L, ZHANG C, ZHANG JY, SUN YP, et al. Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses[J]. Nature Cell Biology, 2022, 24: 353-363. DOI:10.1038/s41556-022-00853-8
|
| [14] |
KUZNETSOV NA, KANAZHEVSKAYA LY, FEDOROVA OS. DNA demethylation in the processes of repair and epigenetic regulation performed by 2-ketoglutarate-dependent DNA dioxygenases[J]. International Journal of Molecular Sciences, 2021, 22(19): 10540. DOI:10.3390/ijms221910540
|
| [15] |
NABEL CS, JIA HJ, YE Y, SHEN L, GOLDSCHMIDT HL, STIVERS JT, ZHANG Y, KOHLI RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation[J]. Nature Chemical Biology, 2012, 8: 751-758. DOI:10.1038/nchembio.1042
|
| [16] |
SALMERÓN-BÁRCENAS EG, ZACAPALA-GÓMEZ AE, TORRES-ROJAS FI, ANTONIO-VÉJAR V, ÁVILA-LÓPEZ PA, BAÑOS-HERNÁNDEZ CJ, NÚÑEZ-MARTÍNEZ HN, DIRCIO-MALDONADO R, MARTÍNEZ-CARRILLO DN, ORTIZ-ORTIZ J, JIMÉNEZ-WENCES H. TET enzymes and 5hmC levels in carcinogenesis and progression of breast cancer: potential therapeutic targets[J]. International Journal of Molecular Sciences, 2023, 25(1): 272. DOI:10.3390/ijms25010272
|
| [17] |
SCHIESSER S, HACKNER B, PFAFFENEDER T, MÜLLER M, HAGEMEIER C, TRUSS M, CARELL T. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing[J]. Angewandte Chemie International Edtion, 2012, 51(26): 6516-6520. DOI:10.1002/anie.201202583
|
| [18] |
LIUTKEVIČIŪTĖ Z, KRIUKIENĖ E, LIČYTĖ J, RUDYTĖ M, URBANAVIČIŪTĖ G, KLIMAŠAUSKAS S. Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases[J]. Journal of the American Chemical Society, 2014, 136(16): 5884-5887. DOI:10.1021/ja5019223
|
| [19] |
SEILER CL, FERNANDEZ J, KOERPERICH Z, ANDERSEN MP, KOTANDENIYA D, NGUYEN ME, SHAM YY, TRETYAKOVA NY. Maintenance DNA methyltransferase activity in the presence of oxidized forms of 5-methylcytosine: structural basis for ten eleven translocation-mediated DNA demethylation[J]. Biochemistry, 2018, 57(42): 6061-6069. DOI:10.1021/acs.biochem.8b00683
|
| [20] |
KUBOSAKI A, TOMARU Y, FURUHATA E, SUZUKI T, SHIN JW, SIMON C, ANDO Y, HASEGAWA R, HAYASHIZAKI Y, SUZUKI H. CpG site-specific alteration of hydroxymethylcytosine to methylcytosine beyond DNA replication[J]. Biochemical and Biophysical Research Communications, 2012, 426(1): 141-147. DOI:10.1016/j.bbrc.2012.08.053
|
| [21] |
LIO CW J, YUE XJ, LÓPEZ-MOYADO IF, TAHILIANI M, ARAVIND L, RAO A. TET methylcytosine oxidases: new insights from a decade of research[J]. Journal of Biosciences, 2020, 45(1): 21. DOI:10.1007/s12038-019-9973-4
|
| [22] |
ARAB K, KARAULANOV E, MUSHEEV M, TRNKA P, SCHÄFER A, GRUMMT I, NIEHRS C. GADD45A binds R-loops and recruits TET1 to CpG island promoters[J]. Nature Genetics, 2019, 51: 217-223. DOI:10.1038/s41588-018-0306-6
|
| [23] |
STOLZ P, MANTERO AS, TVARDOVSKIY A, UGUR E, WANGE LE, MULHOLLAND CB, CHENG YY, WIERER M, ENARD W, SCHNEIDER R, BARTKE T, LEONHARDT H, ELSÄSSER SJ, BULTMANN S. TET1 regulates gene expression and repression of endogenous retroviruses independent of DNA demethylation[J]. Nucleic Acids Research, 2022, 50(15): 8491-8511. DOI:10.1093/nar/gkac642
|
| [24] |
GAO J, MA Y, FU HL, LUO Q, WANG Z, XIAO YH, YANG H, CUI DX, JIN WL. Non-catalytic roles for TET1 protein negatively regulating neuronal differentiation through srGAP3 in neuroblastoma cells[J]. Protein & Cell, 2016, 7(5): 351-361.
|
| [25] |
WU H, D'ALESSIO AC, ITO S, XIA K, WANG ZB, CUI KR, ZHAO KJ, SUN YE, ZHANG Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells[J]. Nature, 2011, 473: 389-393. DOI:10.1038/nature09934
|
| [26] |
曾雨冰, 何学佳, 刘帆, 裴培, 王珊. TET酶的生物学功能及相关机制的研究进展[J]. 生物技术进展, 2022, 12(5): 721-727. ZENG YB, HE XJ, LIU F, PEI P, WANG S. Progress on biological functions and mechanisms of TET enzymes[J]. Current Biotechnology, 2022, 12(5): 721-727 (in Chinese). |
| [27] |
PICCOLO FM, BAGCI H, BROWN KE, LANDEIRA D, SOZA-RIED J, FEYTOUT A, MOOIJMAN D, HAJKOVA P, LEITCH HG, TADA T, KRIAUCIONIS S, DAWLATY MM, JAENISCH R, MERKENSCHLAGER M, FISHER AG. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion[J]. Molecular Cell, 2013, 49(6): 1023-1033. DOI:10.1016/j.molcel.2013.01.032
|
| [28] |
VINCENT JJ, HUANG Y, CHEN PY, FENG SH, CALVOPIÑA JH, NEE K, LEE SA, LE T, YOON AJ, FAULL K, FAN GP, RAO A, JACOBSEN SE, PELLEGRINI M, CLARK AT. Stage-specific roles for Tet1 and Tet2 in DNA demethylation in primordial germ cells[J]. Cell Stem Cell, 2013, 12(4): 470-478. DOI:10.1016/j.stem.2013.01.016
|
| [29] |
ZHANG XC, ZHANG Y, WANG CF, WANG X. TET (ten-eleven translocation) family proteins: structure, biological functions and applications[J]. Signal Transduction and Targeted Therapy, 2023, 8: 297. DOI:10.1038/s41392-023-01537-x
|
| [30] |
CALDWELL BA, BARTOLOMEI MS. DNA methylation reprogramming of genomic imprints in the mammalian germline: a TET-centric view[J]. Andrology, 2023, 11(5): 884-890. DOI:10.1111/andr.13303
|
| [31] |
邱颖, 王红艳. TET去甲基化酶在T细胞中的功能研究[J]. 中国免疫学杂志, 2019, 35(8): 897-901. QIU Y, WANG HY. Function of TET demethylase in T cells[J]. Chinese Journal of Immunology, 2019, 35(8): 897-901 (in Chinese). DOI:10.3969/j.issn.1000-484X.2019.08.001 |
| [32] |
YAMAGUCHI S, SHEN L, LIU YT, SENDLER D, ZHANG Y. Role of Tet1 in erasure of genomic imprinting[J]. Nature, 2013, 504: 460-464. DOI:10.1038/nature12805
|
| [33] |
YAMAGUCHI S, HONG K, LIU R, SHEN L, INOUE A, DIEP D, ZHANG K, ZHANG Y. Tet1 controls meiosis by regulating meiotic gene expression[J]. Nature, 2012, 492: 443-447. DOI:10.1038/nature11709
|
| [34] |
CHRYSANTHOU S, SENNER CE, WOODS L, FINEBERG E, OKKENHAUG H, BURGE S, PEREZ-GARCIA V, HEMBERGER M. A critical role of TET1/2 proteins in cell-cycle progression of trophoblast stem cells[J]. Stem Cell Reports, 2018, 10(4): 1355-1368. DOI:10.1016/j.stemcr.2018.02.014
|
| [35] |
KANG J, LIENHARD M, PASTOR WA, CHAWLA A, NOVOTNY M, TSAGARATOU A, LASKEN RS, THOMPSON EC, SURANI MA, KORALOV SB, KALANTRY S, CHAVEZ L, RAO A. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(31): E4236-E4245.
|
| [36] |
DAI HQ, WANG BG, YANG L, CHEN JJ, ZHU GC, SUN ML, GE H, WANG R, CHAPMAN DL, TANG FC, SUN X, XU GL. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling[J]. Nature, 2016, 538: 528-532. DOI:10.1038/nature20095
|
| [37] |
KHOUEIRY R, SOHNI A, THIENPONT B, LUO XL, VANDE VELDE J, BARTOCCETTI M, BOECKX B, ZWIJSEN A, RAO A, LAMBRECHTS D, KOH KP. Lineage-specific functions of TET1 in the postimplantation mouse embryo[J]. Nature Genetics, 2017, 49: 1061-1072. DOI:10.1038/ng.3868
|
| [38] |
SUN FF, ABREU-RODRIGUEZ I, YE S, GAY S, DISTLER O, NEIDHART M, KAROUZAKIS E. TET1 is an important transcriptional activator of TNFα expression in macrophages[J]. PLoS One, 2019, 14(6): e0218551. DOI:10.1371/journal.pone.0218551
|
| [39] |
LI H, LU T, SUN W, MA RQ, ZHONG H, WEI Y, CHEN DH, WEN YH, CARLSTEN C, WEN WP. Ten-eleven translocation (TET) enzymes modulate the activation of dendritic cells in allergic rhinitis[J]. Frontiers in Immunology, 2019, 10: 2271. DOI:10.3389/fimmu.2019.02271
|
| [40] |
BURLESON JD, SINIARD D, YADAGIRI VK, CHEN XT, WEIRAUCH MT, RUFF BP, BRANDT EB, HERSHEY GKK, JI H. TET1 contributes to allergic airway inflammation and regulates interferon and aryl hydrocarbon receptor signaling pathways in bronchial epithelial cells[J]. Scientific Reports, 2019, 9: 7361. DOI:10.1038/s41598-019-43767-6
|
| [41] |
ISSUREE PD, DAY K, AU C, RAVIRAM R, ZAPPILE P, SKOK JA, XUE HH, MYERS RM, LITTMAN DR. Stage-specific epigenetic regulation of CD4 expression by coordinated enhancer elements during T cell development[J]. Nature Communications, 2018, 9: 3594. DOI:10.1038/s41467-018-05834-w
|
| [42] |
YANG RL, QU CY, ZHOU Y, KONKEL JE, SHI SH, LIU Y, CHEN C, LIU SY, LIU DW, CHEN YB, ZANDI E, CHEN WJ, ZHOU YH, SHI ST. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis[J]. Immunity, 2015, 43(2): 251-263. DOI:10.1016/j.immuni.2015.07.017
|
| [43] |
KRESSLER C, GASPARONI G, NORDSTRÖM K, HAMO D, SALHAB A, DIMITROPOULOS C, TIERLING S, REINKE P, VOLK HD, WALTER J, HAMANN A, POLANSKY JK. Targeted de-methylation of the FOXP3-TSDR is sufficient to induce physiological FOXP3 expression but not a functional Treg phenotype[J]. Frontiers in Immunology, 2021, 11: 609891. DOI:10.3389/fimmu.2020.609891
|
| [44] |
杨晓伟, 赵自亮, 付雨, 于子肖, 赵永聚. TET1基因对小鼠uNK细胞增殖及IFN-γ、VEGF-C和TGF-β1转录水平的影响[J]. 畜牧兽医学报, 2023, 54(3): 1221-1228. YANG XW, ZHAO ZL, FU Y, YU ZX, ZHAO YJ. Effects of TET1 gene on the proliferation of mouse uNK cells and the transcriptional level of IFN-γ, VEGF-C and TGF-β1[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(3): 1221-1228 (in Chinese). |
| [45] |
KOH KP, YABUUCHI A, RAO S, HUANG Y, CUNNIFF K, NARDONE J, LAIHO A, TAHILIANI M, SOMMER CA, MOSTOSLAVSKY G, LAHESMAA R, ORKIN SH, RODIG SJ, DALEY GQ, RAO A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells[J]. Cell Stem Cell, 2011, 8(2): 200-213. DOI:10.1016/j.stem.2011.01.008
|
| [46] |
COSTA Y, DING JJ, THEUNISSEN TW, FAIOLA F, HORE TA, SHLIAHA PV, FIDALGO M, SAUNDERS A, LAWRENCE M, DIETMANN S, DAS S, LEVASSEUR DN, LI Z, XU MJ, REIK W, SILVA JCR, WANG JL. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency[J]. Nature, 2013, 495: 370-374. DOI:10.1038/nature11925
|
| [47] |
ITO S, D'ALESSIO AC, TARANOVA OV, HONG K, SOWERS LC, ZHANG Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification[J]. Nature, 2010, 466(7310): 1129-1133. DOI:10.1038/nature09303
|
| [48] |
LAN J, RAJAN N, BIZET M, PENNING A, SINGH NK, GUALLAR D, CALONNE E, LI GRECI A, BONVIN E, DEPLUS R, HSU PJ, NACHTERGAELE S, MA CJ, SONG RH, FUENTES-IGLESIAS A, HASSABI B, PUTMANS P, MIES F, MENSCHAERT G, WONG JJL, et al. Functional role of Tet-mediated RNA hydroxymethylcytosine in mouse ES cells and during differentiation[J]. Nature Communications, 2020, 11: 4956. DOI:10.1038/s41467-020-18729-6
|
| [49] |
LU FL, LIU YT, JIANG L, YAMAGUCHI S, ZHANG Y. Role of Tet proteins in enhancer activity and telomere elongation[J]. Genes & Development, 2014, 28(19): 2103-2119.
|
| [50] |
YANG J, GUO RP, WANG H, YE XY, ZHOU ZC, DAN JM, WANG HY, GONG P, DENG W, YIN Y, MAO SQ, WANG LB, DING JJ, LI JS, KEEFE DL, DAWLATY MM, WANG JL, XU GL, LIU L. Tet enzymes regulate telomere maintenance and chromosomal stability of mouse ESCs[J]. Cell Reports, 2016, 15(8): 1809-1821. DOI:10.1016/j.celrep.2016.04.058
|
| [51] |
CHRYSANTHOU S, FLORES JC, DAWLATY MM. Tet1 suppresses p21 to ensure proper cell cycle progression in embryonic stem cells[J]. Cells, 2022, 11(8): 1366. DOI:10.3390/cells11081366
|
| [52] |
YANG RL, YU TT, KOU XX, GAO X, CHEN C, LIU DW, ZHOU YH, SHI ST. Tet1 and Tet2 maintain mesenchymal stem cell homeostasis via demethylation of the P2rX7 promoter[J]. Nature Communications, 2018, 9: 2143. DOI:10.1038/s41467-018-04464-6
|
| [53] |
QIAN H, ZHAO JQ, YANG XY, WU SJ, AN Y, QU YX, LI Z, GE H, LI E, QI W. TET1 promotes RXRα expression and adipogenesis through DNA demethylation[J]. Biochimica et Biophysica Acta Molecular and Cell Biology of Lipids, 2021, 1866(6): 158919.
|
| [54] |
DAMAL VILLIVALAM S, YOU D, KIM J, LIM HW, XIAO H, ZUSHIN PJ H, OGURI Y, AMIN PY, KANG S. TET1 is a beige adipocyte-selective epigenetic suppressor of thermogenesis[J]. Nature Communications, 2020, 11: 4313. DOI:10.1038/s41467-020-18054-y
|
| [55] |
ZHANG RR, CUI QY, MURAI K, LIM YC, SMITH ZD, JIN SN, YE P, ROSA L, LEE YK, WU HP, LIU W, XU ZM, YANG L, DING YQ, TANG FC, MEISSNER A, DING CM, SHI YH, XU GL. Tet1 regulates adult hippocampal neurogenesis and cognition[J]. Cell Stem Cell, 2013, 13(2): 237-245. DOI:10.1016/j.stem.2013.05.006
|
| [56] |
CHEN W, LIU NN, SHEN SJ, ZHU W, QIAO J, CHANG SJ, DONG JF, BAI ML, MA L, WANG SS, JIA WW, GUO XD, LI A, XI JJ, JIANG CZ, KANG JH. Fetal growth restriction impairs hippocampal neurogenesis and cognition via Tet1 in offspring[J]. Cell Reports, 2021, 37(5): 109912. DOI:10.1016/j.celrep.2021.109912
|
| [57] |
BAMEZAI S, DEMIR D, PULIKKOTTIL AJ, CICCARONE F, FISCHBEIN E, SINHA A, BORGA C, TE KRONNIE G, MEYER LH, MOHR F, GÖTZE M, CAIAFA P, DEBATIN KM, DÖHNER K, DÖHNER H, GONZÁLEZ-MENÉNDEZ I, QUINTANILLA-FEND L, HEROLD T, JEREMIAS I, FEURING-BUSKE M, et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition[J]. Leukemia, 2021, 35: 389-403. DOI:10.1038/s41375-020-0864-3
|
| [58] |
CIMMINO L, DAWLATY MM, NDIAYE-LOBRY D, YAP YS, BAKOGIANNI S, YU YT, BHATTACHARYYA S, SHAKNOVICH R, GENG HM, LOBRY C, MULLENDERS J, KING B, TRIMARCHI T, ARANDA-ORGILLES B, LIU C, SHEN S, VERMA AK, JAENISCH R, AIFANTIS I. TET1 is a tumor suppressor of hematopoietic malignancy[J]. Nature Immunology, 2015, 16: 653-662. DOI:10.1038/ni.3148
|
| [59] |
HSU CH, PENG KL, KANG ML, CHEN YR, YANG YC, TSAI CH, CHU CS, JENG YM, CHEN YT, LIN FM, HUANG HD, LU YY, TENG YC, LIN ST, LIN RK, TANG FM, LEE SB, HSU HM, YU JC, HSIAO PW, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases[J]. Cell Reports, 2012, 2(3): 568-579. DOI:10.1016/j.celrep.2012.08.030
|
| [60] |
GOOD CR, PANJARIAN S, KELLY AD, MADZO J, PATEL B, JELINEK J, ISSA JP J. TET1-mediated hypomethylation activates oncogenic signaling in triple-negative breast cancer[J]. Cancer Research, 2018, 78(15): 4126-4137. DOI:10.1158/0008-5472.CAN-17-2082
|
| [61] |
PANJARIAN S, ISSA JP J. The roles of DNA demethylases in triple-negative breast cancer[J]. Pharmaceuticals, 2021, 14(7): 628. DOI:10.3390/ph14070628
|
| [62] |
BAO B, TESLOW EA, MITREA C, BOERNER JL, DYSON G, BOLLIG-FISCHER A. Role of TET1 and 5hmC in an obesity-linked pathway driving cancer stem cells in triple-negative breast cancer[J]. Molecular Cancer Research: MCR, 2020, 18(12): 1803-1814. DOI:10.1158/1541-7786.MCR-20-0359
|
| [63] |
LIAO CG, LIANG XH, KE Y, YAO L, LIU M, LIU ZK, HE L, GUO YX, BIAN HJ, CHEN ZN, KONG LM. Active demethylation upregulates CD147 expression promoting non-small cell lung cancer invasion and metastasis[J]. Oncogene, 2022, 41: 1780-1794. DOI:10.1038/s41388-022-02213-0
|
| [64] |
CHEN HQ, CHEN DJ, LI Y, YUAN WB, FAN J, ZHANG Z, HAN F, JIANG X, CHEN JP, WANG DD, CAO J, LIU JY, LIU WB. Epigenetic silencing of TET1 mediated hydroxymethylation of base excision repair pathway during lung carcinogenesis[J]. Environmental Pollution, 2021, 268(Pt B): 115860.
|
| [65] |
MacARTHUR IC, DAWLATY MM. TET enzymes and 5-hydroxymethylcytosine in neural progenitor cell biology and neurodevelopment[J]. Frontiers in Cell and Developmental Biology, 2021, 9: 645335. DOI:10.3389/fcell.2021.645335
|
| [66] |
SAGARKAR S, BHAT N, SAPRE M, DUDHABHATE B, KOKARE DM, SUBHEDAR NK, SAKHARKAR AJ. TET1-induced DNA demethylation in dentate gyrus is important for reward conditioning and reinforcement[J]. Molecular Neurobiology, 2022, 59(9): 5426-5442. DOI:10.1007/s12035-022-02917-0
|
| [67] |
XU WZ, ZHANG XC, LIANG F, CAO YH, LI ZY, QU WZ, ZHANG JY, BI YH, SUN CR, ZHANG JM, SUN BG, SHU Q, LI XK. Tet1 regulates astrocyte development and cognition of mice through modulating GluA1[J]. Frontiers in Cell and Developmental Biology, 2021, 9: 644375. DOI:10.3389/fcell.2021.644375
|
| [68] |
ZHOU DX, HUANG Z, ZHU XX, HONG T, ZHAO YL. Circular RNA 0025984 ameliorates ischemic stroke injury and protects astrocytes through miR-143-3p/TET1/ORP150 pathway[J]. Molecular Neurobiology, 2021, 58(11): 5937-5953. DOI:10.1007/s12035-021-02486-8
|
| [69] |
MOYON S, FRAWLEY R, MARECHAL D, HUANG D, MARSHALL-PHELPS KLH, KEGEL L, BØSTRAND SMK, SADOWSKI B, JIANG YH, LYONS DA, MÖBIUS W, CASACCIA P. TET1-mediated DNA hydroxymethylation regulates adult remyelination in mice[J]. Nature Communications, 2021, 12: 3359. DOI:10.1038/s41467-021-23735-3
|
| [70] |
ZHANG M, WANG J, ZHANG KX, LU GZ, LIU YM, REN KK, WANG WT, XIN DZ, XU LL, MAO HH, XING JL, GAO XC, JIN WL, BERRY K, MIKOSHIBA K, WU SX, LU QR, ZHAO XH. Ten-eleven translocation 1 mediated-DNA hydroxymethylation is required for myelination and remyelination in the mouse brain[J]. Nature Communications, 2021, 12: 5091. DOI:10.1038/s41467-021-25353-5
|
| [71] |
GREER CB, WRIGHT J, WEISS JD, LAZARENKO RM, MORAN SP, ZHU J, CHRONISTER KS, JIN AY, KENNEDY AJ, SWEATT JD, KAAS GA. Tet1 isoforms differentially regulate gene expression, synaptic transmission, and memory in the mammalian brain[J]. The Journal of Neuroscience, 2021, 41(4): 578-593. DOI:10.1523/JNEUROSCI.1821-20.2020
|
| [72] |
WANG B, ZHAO T, CHEN XX, ZHU YY, LU X, QIAN QH, CHEN HR, MENG XH, WANG H, WEI W, XU DX. Gestational 1-nitropyrene exposure causes anxiety-like behavior partially by altering hippocampal epigenetic reprogramming of synaptic plasticity in male adult offspring[J]. Journal of Hazardous Materials, 2023, 453: 131427. DOI:10.1016/j.jhazmat.2023.131427
|
| [73] |
XU T, STEWART KM, WANG XH, LIU K, XIE M, RYU JK, LI K, MA TH, WANG HX, NI L, ZHU SY, CAO N, ZHU DW, ZHANG Y, AKASSOGLOU K, DONG C, DRIGGERS EM, DING S. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism[J]. Nature, 2017, 548(7666): 228-233. DOI:10.1038/nature23475
|
| [74] |
CHONG TL, AHEARN EL, CIMMINO L. Reprogramming the epigenome with vitamin C[J]. Frontiers in Cell and Developmental Biology, 2019, 7: 128. DOI:10.3389/fcell.2019.00128
|
| [75] |
SHEKHAWAT J, GAUBA K, GUPTA S, CHOUDHURY B, PUROHIT P, SHARMA P, BANERJEE M. Ten-eleven translocase: key regulator of the methylation landscape in cancer[J]. Journal of Cancer Research and Clinical Oncology, 2021, 147(7): 1869-1879. DOI:10.1007/s00432-021-03641-3
|
| [76] |
MINOR EA, COURT BL, YOUNG JI, WANG GF. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine[J]. The Journal of Biological Chemistry, 2013, 288(19): 13669-13674. DOI:10.1074/jbc.C113.464800
|
| [77] |
YIN RC, MAO SQ, ZHAO BL, CHONG ZC, YANG Y, ZHAO C, ZHANG DP, HUANG H, GAO J, LI Z, JIAO Y, LI CP, LIU SQ, WU DN, GU WK, YANG YG, XU GL, WANG HL. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals[J]. Journal of the American Chemical Society, 2013, 135(28): 10396-10403. DOI:10.1021/ja4028346
|
| [78] |
JAKUBEK M, KEJÍK Z, KAPLÁNEK R, ANTONYOVÁ V, HROMÁDKA R, ŠANDRIKOVÁ V, SÝKORA D, MARTÁSEK P, KRÁL V. Hydrazones as novel epigenetic modulators: correlation between TET 1 protein inhibition activity and their iron(Ⅱ) binding ability[J]. Bioorganic Chemistry, 2019, 88: 102809. DOI:10.1016/j.bioorg.2019.02.034
|
| [79] |
TSAI YP, CHEN HF, CHEN SY, CHENG WC, WANG HW, SHEN ZJ, SONG CX, TENG SC, HE C, WU KJ. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator[J]. Genome Biology, 2014, 15(12): 513.
|
| [80] |
ALI MM, PHILLIPS SA, MAHMOUD AM. HIF1α/TET1 pathway mediates hypoxia-induced adipocytokine promoter hypomethylation in human adipocytes[J]. Cells, 2020, 9(1): 134.
|