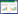中国科学院微生物研究所、中国微生物学会主办
文章信息
- 柴文婷, 杨博慧, 赵珊珊, 郭志强, 朱立勋, 范佳利, 杨伟, 赵威军, 郝艳平, 吕晋慧, 孙文献, 张春来
- CHAI Wenting, YANG Bohui, ZHAO Shanshan, GUO Zhiqiang, ZHU Lixun, FAN Jiali, YANG Wei, ZHAO Weijun, HAO Yanping, LÜ Jinhui, SUN Wenxian, ZHANG Chunlai
- 高粱MYC基因家族序列、表达模式及自然等位变异分析
- Characterization of sequences, expression profiling, and natural allelic variation analysis of the MYC gene family in sorghum (Sorghum bicolor)
- 生物工程学报, 2024, 40(4): 1170-1194
- Chinese Journal of Biotechnology, 2024, 40(4): 1170-1194
- 10.13345/j.cjb.230641
-
文章历史
- Received: September 18, 2023
- Accepted: November 28, 2023
- Published: December 4, 2023
2. 山西农业大学高粱研究所, 山西 榆次 030600;
3. 中国农业大学植物保护学院, 北京 100193
2. Institute for Sorghum Research, Shanxi Agricultural University, Yuci 030600, Shanxi, China;
3. College of Plant Protection, China Agricultural University, Beijing 100193, China
高粱是我国重要的杂粮作物之一,广泛应用于酿造、饲料和生物能源等多个产业[1]。高粱蚜和丝黑穗病分别是高粱主产区的主要虫害和病害,其发生和流行使产量和品质下降。研究表明MYC转录因子可调节茉莉酸的合成,诱导相关抗性基因表达,一些MYC成员可负调控脯氨酸生物合成,参与调控植物抗逆性。拟南芥MYC2、MYC3和MYC4参与调节种子贮藏蛋白的积累,表明MYC转录因子在提高抗逆性、产量和品质方面有较大潜力[2-3]。
MYC基因在调节植物生长发育和次生代谢产物积累上发挥重要作用[4]。目前已经鉴定多个MYC基因,包括水稻7个、玉米7个、小麦26个、二穗短柄草7个和红豆杉5个[5-7]。Ludwig等[8]发现玉米R基因产物Lc是植物中首个含基本螺旋-环-螺旋(basic helix-loop-helix, bHLH)结构域的蛋白,参与黄酮和花青素合成。Hichri等[9]发现葡萄VvMYC1和VvMYBA1诱导花青素或原花青素(proanthocyanidin, PA)合成。MYC2在蓝光介导下调节拟南芥生长发育过程中的基因表达,遗传分析发现AtMYC2参与蓝光介导的光形态生长,是蓝光和远红光调节基因表达的负调节因子[10-11],在盐胁迫条件下,MYC2受丝裂原活化蛋白激酶(mitogen-activated protein kinase, MAPK)激活,调节脯氨酸的生物合成,从而调节盐胁迫反应[12]。AtMYC3和AtMYC4均能调节植株合成花青素[13-15],红豆杉MYC基因可参与调控次生代谢产物紫杉醇的合成[16]。作为bHLH的亚族,MYC基因家族含有bHLH_MYC_N结构域,这是蛋白质相互作用所必需的,HLH结构域可以促进蛋白质与DNA结合[17]。
高粱植株和籽粒可积累花青素、单宁等次生代谢物,控制籽粒颜色的基因分别为Y、R、Z和I,分别位于SBI-01、SBI-03、SBI-02和SBI-07染色体上,Y和R分别编码MYB (Sobic.001G397900)和DFR (Sobic.003G230900、Sobic.003G231000)[18-19],冯周等[20]定位了Z,初步分析发现其候选基因Sobic.002G204200编码泛素连接酶,还未鉴定到I的候选基因。控制籽粒单宁含量的2个主效位点Tan1 (Sorbic.004G280800)和Tan2 (Sorbic.002G076600)分别位于SBI-04和SBI-02,编码WD40和MYC[21-22]。Lv等[23]定位了SBI-07上一个红叶基因RL (Sobic.007G234100)编码细胞壁关联的激酶。高粱生长过程中受干旱、盐碱和病虫等胁迫,导致减产。全基因组关联分析(genome-wide association analysis, GWAS)关联到SBI-06上控制花青素积累、抗冷的MYC基因[24];表皮蜡含量与SBI-03上的bHLH13关联[25],用分子生物学方法也分离到数个控制抗盐的bHLH[26-27]。
现代农业的绿色发展要求对高粱品种选育和病虫害防治提出新挑战。高粱蚜(Melanaphis sacchari)通过刺吸式口器从高粱组织获取营养,造成叶片失绿,严重时穗不能抽出,籽粒败育,是高粱生产上重要的虫害[28]。丝轴黑粉菌(Sporisorium reilianum)侵染高粱幼苗,导致抽穗期形成花序充满菌丝和孢子,不能结实,是高粱生产上毁灭性的病害[29]。生产上主要通过培育抗蚜、抗病品种进行防治。国内外通过遗传作图和基因组关联分析,初步定位抗丝黑穗病和抗蚜基因[30]。
高粱全基因组DNA的测序工作在2009年初步完成,于2018年得到完善[31],这使得从基因组水平来揭示高粱重要基因家族的功能成为可能。目前关于高粱MYC基因的研究较少,本研究对MYC基因表达模式及响应高粱蚜、丝轴黑粉菌侵染与自然等位DNA变异分析等进行了研究,旨在为研究MYC的生物学功能提供理论依据。
1 材料与方法 1.1 高粱MYC家族成员基因鉴定、染色体定位及共线性分析以拟南芥(Arabidopsis thaliana)和水稻(Oryza sativa) MYC氨基酸序列作为参考,在NCBI数据库(http://www.ncbi.nlm.nih.gov/)中进行BLASTP搜寻和比对,利用CDD和Pfam筛选出含有该结构域的蛋白质序列。在Phytozome数据库(V12, https://phytozome.jgi.doe.gov/pz/portal.html)下载高粱的基因组注释文件以及SbMYCs基因序列和氨基酸序列。SbMYCs编号以NCBI命名为主,少数结合了已完成的功能分析的MYC基因[32]。同时获取玉米(Zea mays)、大豆(Glycine max)、水稻和拟南芥的蛋白序列,采用TBtools[33]软件进行高粱MYC基因染色体定位及共线性信息绘图。
1.2 高粱MYC转录因子家族成员基因结构分析利用MEGA 11.0软件的邻接法(neighbor- joining, NJ)构建系统进化树[34-35],利用GSDS在线工具(Gene Structure Display Server 2.0, http://gsds.cbi.pku.edu.cn/)绘制高粱MYC基因结构图。通过MEME[36]获取SbMYC家族motif位置分布信息,保守序列的最大鉴定数目设置为10。
1.3 高粱MYC基因家族蛋白质理化性质分析从Phytozome数据库和ExPASy-Compute pI/Mw (http://web.expasy.org/compute_pi/)[37]获取SbMYC蛋白理化性质相关信息,包括氨基酸数目、分子量大小、理论等电点、不稳定系数、脂溶性系数和亲水性平均值。
1.4 高粱MYC家族蛋白二级结构、亚细胞定位分析采用PSORT (psort1.hgc.jp/form.html)和SOPMA[38] (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)分别对高粱MYC蛋白二级结构和亚细胞定位进行分析。
1.5 高粱MYC基因启动子顺式作用元件分析从NCBI下载MYC基因启动子上游2 000 bp序列,提交至PlantCARE数据库(Plant Cis-Acting Regulatory Element, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/),进行SbMYCs的启动子顺式作用元件分析。
1.6 高粱MYC系统进化树分析通过MEGA 11.0软件,对高粱、拟南芥、水稻、玉米和大豆MYC家族氨基酸序列进行比对,用邻接法构建系统进化树,采用iTOL (http://itol.embl.de)在线工具对进化树进行美化。
1.7 高粱MYC家族进化选择压力分析通过MEGA 11.0软件对SbMYC家族基因的编码序列(coding sequence, CDS)进行对比,再利用Tbtools软件中Simplement Ka/Ks Calculator (NG)计算出基因的进化选择压力。
1.8 高粱MYC基因表达分析从Phytozome数据库中下载高粱MYC基因在各个生育期、不同器官部位的表达量,利用TBtools软件作出表达量的热图。
1.9 SbMYC基因表达及蚜虫和丝黑穗侵染影响分析抗、感高粱蚜品系2457B、5-27sugB在可控光照生长箱生长,抗丝黑穗病品系1383-2×7050B F4-12Res、感病品系1383-2×7050B F4-12Sus以及R111和SSA1在温室生长,高粱丝轴黑粉菌、高粱蚜侵染取样及基因表达分析的详细步骤参照赵珊珊等[39]的方法。在授粉后10 d和20 d,对幼嫩籽粒取样,提取RNA,采用实时荧光定量PCR (quantitative real-time PCR detecting system, qPCR)检测基因表达水平。qPCR参照范佳利等[40]的方法,基因专一性引物见表 1。
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
| SbMYC2.1g | TGCCATCTTTTGGCAGTCCT | GCCGGAGATGAGGGAATTGA |
| SbLHW.2g | TGGATGCATGGGAATCAACA | CATCGATGCTTTGATGCATTTGG |
| SbTan2/GLABRA3 | GTTCTACCTCATGTGCGCCT | GGACACCATCGTTGACTGGA |
| SbMYC2.2g2 | GCTGTACGTGTGAGGAGCTA | ATGACGGCTCGGAAAAGGAG |
| SbMYC2.2g1 | TGACAATGGACGACCTGCTC | CTTCCCACTGGCCATAGGTC |
| SbbHLH13.3g | AGCTCTTGTTGAATCCATTGAGG | GGCATAGTAACAAGATGAACATGGC |
| SbMYC2.3g | CAACGGTGCAATGGATGACC | CTGGTGGAGCTCATGTCCTG |
| SbAbaIn | ATCTTCGGCAAGGACCTCTC | GAAGTCTATGCTCCTGGGCG |
| SbbHLH155.3g | GAAGGGCTGATGGGAAAGGT | ATGGTCTGGATGCCTGATGC |
| SbTDR | TAGGTGACCTGCCTCCATCA | GTGGTCTAGGAGCACGTCTG |
| SbbHLH155.4g | CGGCATATACGACCGGACAA | TGAAGAAGAGCTCAGGCTGC |
| SbLHW.4g | ATATGGGGAGAAACCGCCAC | ACTCGCCTGTCCTGAAACAG |
| SbLHW.5g | AGCCTGGTCCATTTGGAAGC | CTGCTGGGTCTGGAACTCTG |
| SbMYC4.5g | TGCCATCTTTTGGCAGTCCT | GCCGGAGATGAGGGAATTGA |
| SbR-S4 | GGTCCTACTCCGATGAAGCG | TTTCCTGGGCTGTTCGTACC |
| SbR-S3 | ATACTCGGAAGAGCCGACCT | GCATTGAACAAGCTGACGGG |
| SbR-S2 | CAGGAATAGTAAGGCGAGCG | TGTAGAACCCGTCCGTCCAT |
| SbR-S1 | CCTTCCGCCTCAACAGGAAT | CGACCTGGTTGAGTGCTTGA |
| SbLHW.6g | ATATTTATCAGCCCCCTCTCCC | AGGGTCTTGGGGATGGAAGC |
| SbbHLH35.7g | GCCCCTTGCGCATTATAGGA | TCCCCTTGTAGAGACCATCC |
| SbMYC2.7g1 | GTCCCCCAGTCATCCTTCTG | TTCTTCGCCGAGTAGCCATT |
| SbMYC4.7g | TCAGTCCCCAGCCTTCTTCT | GTGCCTAACACTGTCGCTGT |
| SbMYC2.7g2 | AACGCTTGAGCTGGATCACT | AACATCAGTGGAGACGCAGT |
| SbMYC3.7g | TGCCCTGTGATTTCCTGGAG | GGCTGTTGGTGTTGTTACGG |
| SbLHW.8g | CTGAAGAAGCTGGGTGTGAG | AACCTCGGGTCTGAGATTGTG |
| SbMYC2.9g | GCCATTCTCGGCTTGCACTA | GCATCTTTGCAGCTGATGCT |
| SbbHLH13.9g | TGACCAGACAGAAGATGCGG | CACCAACGTGACGTGGAAAC |
| SbbHLH155.10g | CATATGCCGGGGACTGTGAG | GCCGCCACTTTCCCTATCAG |
| 18S rRNA | GCCTTTCGAAGCACTTTCAC | AACCAAACCTCCATGCTCAC |
对上述高粱基因型以及1383-2/15、3030、2457A3、91SPbigspike、B35、R111、Apo111、3chi3Mid、Pin626、JF1、4003、961541、Xinliang7、EBA-3、C1-1-1、44B、Jinliang5、SP42、V4A2、E35B、SP91dw和961541H在BMKcloud.com平台分析DNA变异,获得SbMYC基因单核苷酸多态性(single nucleotide polymorphism, SNP)、插入缺失标记(insertion-deletion, INDEL)变异位点,通过Excel筛选整理整合相同基因型,统计变异位点,分析单倍型。其中2457A3、V4A2为高粱杂交种组配的雄性不育系,2457B、5-27sugB和44B为雄性不育系的保持系,1383-2/15、3030、91SPbigspike、R111、Apo111、3chi3Mid、Pin626、JF1、4003、961541、Xinliang7、C1-1-1、Jinliang5、SP42、E35B、SP91dw和961541H为雄性不育系的恢复系,B35为叶片持绿特异种质,EBA-3为含Ma5和Ma6的特异种质,均由本团队收集和保存。
1.11 高粱SbTan2基因单倍型分析根据高粱单宁含量测定方法(GB/T 15686―2008)对上述高粱材料单宁含量进行测定,利用上述重测序中的DNA变异信息,分析单倍型和表型数据的关联性。
1.12 SbMYC互作蛋白预测通过STRING数据库(https://cn.string-db.org/),交互蛋白数量设置为10个以内,构建高粱MYC互作蛋白网络图。
2 结果与分析 2.1 SbMYCs基因家族基本信息、染色体定位及共线性分析结合高粱转录组测序数据,并通过结构域验证最终获取到28个MYC基因(表 2)。染色体定位发现MYC基因在高粱10条染色体上均有分布,SBI-06和SBI-07号染色体上基因个数最多(5个),SBI-01、SBI-08和SBI-10号染色体上分布基因最少(仅1个),多数SbMYCs基因分布在染色体两端(图 1),可能避免了异染色质区,基因长度为771−17 879 bp,CDS序列291−2 676 bp。
| Gene name | Gene ID | Chromosome | Genome location | Length (bp) | CDS (bp) |
| SbMYC2.1g | Sobic.001G287600 | SBI-01 | 56 318 190−56 321 348 | 3 158 | 2 124 |
| SbLHW.2g | Sobic.002G001500 | SBI-02 | 196 754−57 321 348 | 5 405 | 1 908 |
| SbTan2/GLABRA3[32] | Sobic.002G076600 | SBI-02 | 7 975 936−58 321 348 | 9 285 | 2 034 |
| SbMYC2.2g2 | Sobic.002G267000 | SBI-02 | 65 126 279−59 321 348 | 1 098 | 831 |
| SbMYC2.2g1 | Sobic.002G267100 | SBI-02 | 65 136 135−60 321 348 | 1 443 | 891 |
| SbbHLH13.3g | Sobic.003G004500 | SBI-03 | 429 258−61 321 348 | 3 564 | 1 869 |
| SbMYC2.3g | Sobic.003G077100 | SBI-03 | 656 034−62 321 348 | 771 | 771 |
| SbAbaIn | Sobic.003G272200 | SBI-03 | 60 842 727−63 321 348 | 2 434 | 1 455 |
| SbbHLH155.3g | Sobic.003G324600 | SBI-03 | 65 124 587−64 321 348 | 4 815 | 1 380 |
| SbTDR | Sobic.004G017500 | SBI-04 | 1 370 525−65 321 348 | 3 478 | 1 716 |
| SbbHLH155.4g | Sobic.004G241500 | SBI-04 | 58 961 101−66 321 348 | 3 283 | 1 149 |
| SbLHW.4g | Sobic.004G284600 | SBI-04 | 62 680 454−67 321 348 | 5 063 | 2 334 |
| SbLHW.5g | Sobic.005G045100 | SBI-05 | 4 268 573−68 321 348 | 6 145 | 2 613 |
| SbMYC4.5g | Sobic.005G061601 | SBI-05 | 671 436−69 321 348 | 999 | 219 |
| SbR-S4 | Sobic.006G076900 | SBI-06 | 44 126 382−70 321 348 | 14 007 | 1 770 |
| SbR-S3 | Sobic.006G175200 | SBI-06 | 53 044 178−71 321 348 | 5 199 | 1 773 |
| SbR-S2 | Sobic.006G175500 | SBI-06 | 53 062 306−72 321 348 | 17 878 | 1 641 |
| SbR-S1 | Sobic.006G175700 | SBI-06 | 53 102 700−73 321 348 | 8 329 | 1 767 |
| SbLHW.6g | Sobic.006G182000 | SBI-06 | 53 688 464−74 321 348 | 5 392 | 2 517 |
| SbbHLH35.7g | Sobic.007G051800 | SBI-07 | 5 299 966−75 321 348 | 2 088 | 867 |
| SbMYC2.7g1 | Sobic.007G182900 | SBI-07 | 61 594 875−76 321 348 | 916 | 837 |
| SbMYC4.7g | Sobic.007G183000 | SBI-07 | 61 602 262−77 321 348 | 840 | 840 |
| SbMYC2.7g2 | Sobic.007G183050 | SBI-07 | 61 608 734−78 321 348 | 792 | 792 |
| SbMYC3.7g | Sobic.007G183200 | SBI-07 | 61 641 933−79 321 348 | 4 885 | 813 |
| SbLHW.8g | Sobic.008G044000 | SBI-08 | 4 344 861−80 321 348 | 5 783 | 2 676 |
| SbMYC2.9g | Sobic.009G036333 | SBI-09 | 3 328 869−81 321 348 | 3 975 | 1 263 |
| SbbHLH13.9g | Sobic.009G088100 | SBI-09 | 15 112 562−82 321 348 | 4 235 | 1 764 |
| SbbHLH155.10g | Sobic.010G095800 | SBI-10 | 8 617 650−83 321 348 | 3 037 | 1 203 |
|
| 图 1 SbMYCs基因家族染色体分布 Fig. 1 Chromosome location of SbMYCs. |
| |
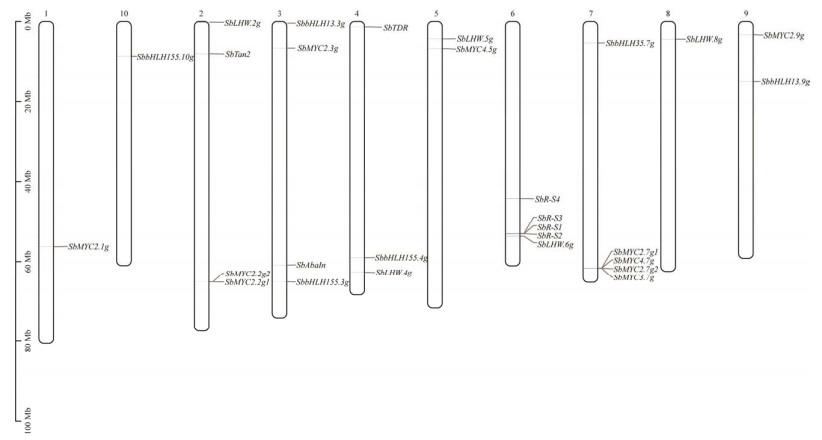
对SbMYC基因家族成员与拟南芥和水稻进行共线性分析(图 2),结果显示,SbMYCs与拟南芥存在3个同源基因对,与水稻有21个同源基因对,表明高粱和水稻MYC基因功能更相近,调控途径更相似。
|
| 图 2 SbMYCs基因共线性分析 Fig. 2 Collinear analysis of SbMYCs. |
| |
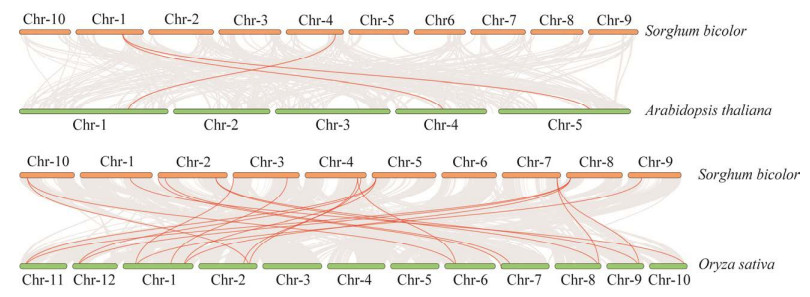
如图 3A所示,将SbMYCs基因进行聚类分析,共分为5个亚族,同一亚族下亲缘关系越近,motif分布的种类、数量、位置、内含子-外显子结构和保守基序越相似。
|
| 图 3 SbMYCs基因聚类图(A)、基因结构(B)、结构域(C)和motif位置分布(D) Fig. 3 SbMYCs cluster diagram (A), gene structure (B), domain (C) and motif location distribution (D). |
| |
高粱MYC基因结构分析见图 3B,SbMYCs基因内含子数量的变化范围从0−11个不等。除SbMYC2.2g2、SbMYC2.7g1、SbMYC2.3g、SbMYC2.7g2、SbMYC4.7g、SbR-S3和SbTan2这8个基因上下游非编码区有缺失,其余SbMYC基因均存在上下游非编码区。其中SbLHW.4g、SbLHW.6g、SbTan2/GLABRA3、SbLHW.2g、SbLHW.5g和SbLHW.8g的内含子数量最多,高达11个;多数Ⅰ类成员不含内含子。
对高粱MYC蛋白保守结构域进一步分析(图 3C)发现,高粱MYC蛋白包含HLH和bHLH_MYC_N两种结构域,其中8个MYC转录因子同时包含这2种结构域;18个MYC蛋白含有保守结构域bHLH_MYC_N。
保守基序有助于理解MYC转录因子的功能,对高粱MYC转录因子motif位置分布进行分析(图 3D),共发现10个motif。其中motif 1、motif 3和motif 4均有50个氨基酸(图 4)。Motif 1和motif 2分布数量最多,分布在25个和27个高粱MYC蛋白中,SbMYC4.5g仅存在motif 4。说明motif 1和motif 2为较为保守的基序,与MYC基因的功能密切相关。

|
| 图 4 SbMYCs蛋白10个保守基序序列标识 Fig. 4 Sequence logos of the 10 conserved motifs in SbMYCs proteins. |
| |
由表 3可知:SbMYC蛋白质编码氨基酸数目在72−891 aa之间;分子量大小在7 659.79−94 767.82 Da之间;理论等电点在4.51−7.15之间;25个SbMYC蛋白质等电点小于7为酸性蛋白。不稳定系数分析表明,26个SbMYC蛋白为不稳定蛋白,SbLHW.4g和SbMYC4.5g为稳定蛋白。SbMYC脂溶性系数在54.77−96.25之间,除SbLHW.2g和SbMYC4.5g蛋白外,其余26个SbMYC蛋白的脂溶系数均小于90,表明SbMYC蛋白的流动性较弱。亲水性平均值都小于0,表明SbMYC蛋白质均为亲水性蛋白。
| Proteins | Number of amino acids (aa) | Molecular weight (Da) | Theoretical pI | Instability index | Aliphatic index | Average hydrophilicity |
| SbMYC2.1g | 707 | 75 519.31 | 6.12 | 54.72 | 63.42 | −0.524 |
| SbLHW.2g | 635 | 71 020.99 | 5.47 | 50.67 | 90.43 | −0.232 |
| SbTan2/GLABRA3 | 677 | 73 741.47 | 6.16 | 58.86 | 78.40 | −0.449 |
| SbMYC2.2g2 | 276 | 29 255.02 | 5.74 | 62.37 | 85.65 | −0.287 |
| SbMYC2.2g1 | 296 | 31 907.83 | 5.21 | 67.03 | 76.22 | −0.435 |
| SbbHLH13.3g | 622 | 67 721.55 | 7.12 | 47.20 | 75.43 | −0.466 |
| SbMYC2.3g | 256 | 27 582.10 | 6.05 | 48.53 | 86.21 | −0.297 |
| SbAbaIn | 484 | 51 219.49 | 7.26 | 48.48 | 73.16 | −0.388 |
| SbbHLH155.3g | 459 | 49 025.83 | 5.97 | 54.25 | 54.77 | −0.506 |
| SbTDR | 571 | 59 313.95 | 4.53 | 49.04 | 77.79 | −0.251 |
| SbbHLH155.4g | 382 | 40 208.15 | 5.25 | 48.56 | 75.84 | −0.076 |
| SbLHW.4g | 777 | 84 906.83 | 5.41 | 40.00 | 80.55 | −0.286 |
| SbLHW.5g | 870 | 94 767.82 | 6.29 | 45.98 | 78.56 | −0.369 |
| SbMYC4.5g | 72 | 7 659.79 | 4.51 | 38.49 | 96.25 | 0.369 |
| SbR-S4 | 587 | 64 424.19 | 5.93 | 45.43 | 81.11 | −0.324 |
| SbR-S3 | 580 | 63 187.17 | 5.03 | 49.14 | 81.26 | −0.305 |
| SbR-S2 | 546 | 59 640.43 | 6.04 | 62.40 | 79.16 | −0.469 |
| SbR-S1 | 588 | 65 219.53 | 5.29 | 50.00 | 75.85 | −0.409 |
| SbLHW.6g | 824 | 91 047.04 | 5.14 | 48.13 | 79.16 | −0.457 |
| SbbHLH35.7g | 288 | 31 252.14 | 4.72 | 66.75 | 82.43 | −0.320 |
| SbMYC2.7g1 | 278 | 29 532.26 | 6.31 | 65.69 | 77.30 | −0.319 |
| SbMYC4.7g | 279 | 29 750.77 | 6.53 | 55.20 | 83.30 | −0.193 |
| SbMYC2.7g2 | 263 | 28 104.60 | 6.06 | 51.92 | 79.58 | −0.323 |
| SbMYC3.7g | 270 | 28 987.88 | 7.15 | 53.78 | 85.41 | −0.227 |
| SbLHW.8g | 891 | 96 916.30 | 6.64 | 42.11 | 74.76 | −0.437 |
| SbMYC2.9g | 420 | 43 577.55 | 5.04 | 73.64 | 72.21 | −0.330 |
| SbbHLH13.9g | 587 | 64 288.53 | 6.19 | 46.91 | 79.42 | −0.426 |
| SbbHLH155.10g | 400 | 42 077.11 | 4.96 | 60.39 | 70.17 | −0.242 |
高粱MYC家族成员蛋白的二级结构预测结果见表 4,高粱MYC家族各成员的二级结构中均含有α-螺旋、β-折叠、延伸链和无规则卷曲。其中,α-螺旋(25.93%−51.48%)和无规则卷曲(37.78%−54.68%)为二级结构的主要存在形式,而延伸链和β-折叠占比则相对较少,散布在整个蛋白质中。
| Proteins | Alpha-helix (%) | Βeta-turn (%) | Extension chain (%) | Random coil (%) | Intracellular location | Probability (%) |
| SbMYC2.1g | 31.82 | 2.55 | 11.32 | 54.31 | Nucleus | 70.0 |
| SbLHW.2g | 41.42 | 3.62 | 12.44 | 42.52 | Nucleus | 76.0 |
| SbTan2/GLABRA3 | 37.22 | 2.81 | 9.45 | 50.52 | Nucleus | 30.0 |
| SbMYC2.2g2 | 40.58 | 1.81 | 11.59 | 46.01 | Chloroplast stroma | 52.4 |
| SbMYC2.2g1 | 42.23 | 3.04 | 10.81 | 43.92 | Chloroplast stroma | 57.7 |
| SbbHLH13.3g | 33.60 | 3.70 | 13.02 | 49.68 | Nucleus | 70.0 |
| SbMYC2.3g | 36.92 | 4.64 | 13.92 | 44.51 | Cytoplasm | 45.0 |
| SbAbaIn | 36.57 | 3.10 | 11.98 | 48.35 | Endoplasmic reticulum | 55.0 |
| SbbHLH155.3g | 25.93 | 5.66 | 13.73 | 54.68 | Microbody | 69.4 |
| SbTDR | 38.92 | 6.11 | 13.26 | 41.71 | Nucleus | 98.0 |
| SbbHLH155.4g | 41.36 | 7.33 | 10.73 | 40.58 | Microbody | 52.2 |
| SbLHW.4g | 31.92 | 6.69 | 17.63 | 43.76 | Microbody | 76.0 |
| SbLHW.5g | 30.46 | 5.40 | 12.64 | 51.49 | Nucleus | 76.0 |
| SbMYC4.5g | 26.39 | 8.33 | 23.61 | 41.67 | Microbody | 64.0 |
| SbR-S4 | 41.57 | 4.43 | 11.07 | 42.93 | Plasma membrane | 79.0 |
| SbR-S3 | 40.17 | 3.28 | 11.03 | 45.52 | Plasma membrane | 79.0 |
| SbR-S2 | 37.00 | 3.11 | 12.82 | 47.07 | Nucleus | 96.0 |
| SbR-S1 | 41.67 | 3.40 | 11.90 | 43.03 | plasma membrane | 60.0 |
| SbLHW.6g | 28.88 | 4.61 | 15.05 | 51.46 | Nucleus | 76.0 |
| SbbHLH35.7g | 46.88 | 0.00 | 11.11 | 39.24 | Chloroplast thylakoid space | 80.0 |
| SbMYC2.7g1 | 50.00 | 2.88 | 8.63 | 38.49 | Chloroplast stroma | 84.7 |
| SbMYC4.7g | 46.59 | 1.79 | 9.32 | 42.29 | Chloroplast stroma | 90.4 |
| SbMYC2.7g2 | 48.29 | 2.28 | 7.98 | 41.44 | Chloroplast stroma | 52.6 |
| SbMYC3.7g | 51.48 | 2.59 | 8.15 | 37.78 | Chloroplast stroma | 51.6 |
| SbLHW.8g | 28.28 | 5.84 | 14.59 | 51.29 | Nucleus | 76.0 |
| SbMYC2.9g | 44.76 | 4.29 | 8.10 | 42.86 | Endoplasmic reticulum | 85.0 |
| SbbHLH13.9g | 37.31 | 3.41 | 12.10 | 47.19 | Microbody | 30.0 |
| SbbHLH155.10g | 41.50 | 7.00 | 11.75 | 39.75 | Nucleus | 76.0 |
对高粱MYC家族成员进行亚细胞定位,结果表明,28个MYC蛋白分别定位于细胞核、叶绿体、细胞质、内质网、微体和质膜内,SbMYC2.1g、SbLHW.2g等10个蛋白定位于细胞核,概率区间为30%−98%,SbMYC2.2g1、SbMYC2.2g2等7个定位于叶绿体基质,概率区间为51.6%−90.4%。SbLHW.4g、SbMYC4.5g和SbbHLH155.4g等5个定位于微体,SbAbaIn和SbMYC2.9g定位于内质网。
2.5 高粱MYC基因启动子分析高粱MYC家族基因启动子顺式作用元件分析(图 5)表明,SbMYC基因家族各成员顺式作用元件的种类和数量都有差异。在MYC基因启动子中发现了与植物生长发育、激素调控和胁迫响应相关等不同类型的顺式元件。其中23个SbMYC成员启动子区域含有顺式元件ABRE,参与脱落酸反应;并发现与光调控相关的元件G-box、CGTCA-motif和TGACG-motif存在于22个启动子中,与茉莉酸甲酯(methyl jasmonate, MeJA)诱导植物防御反应相关;22个启动子中存在抗氧化逆境响应元件ARE,与厌氧诱导相关。
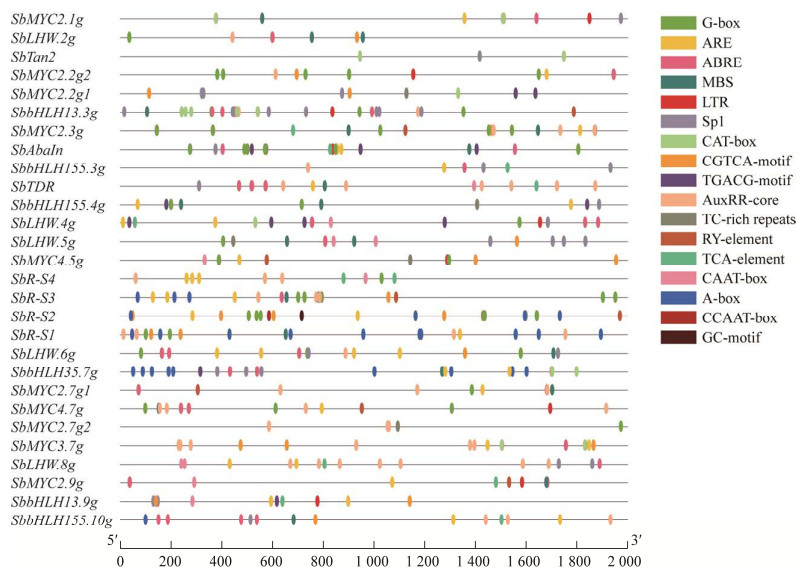
|
| 图 5 SbMYCs基因启动子区的顺式作用调控元件核心序列及功能 Fig. 5 Core sequences and function of the cis-acting regulatory elements in promoter regions of SbMYCs. |
| |
此外,与光反应(Sp1、G-Box)、水杨酸(TCA-element)、分生组织表达(CAT-box)、生长素反应(AuxRR-core)、种子调控(RY-element)相关的顺式元件,以及与响应逆境胁迫相关的干旱诱导元件MBS、低温响应元件LTR和防御、胁迫响应元件TC-rich repeats等顺势元件均存在于SbMYCs启动子中。顺式元件分析表明,SbMYCs在植物的生长发育、信号转导、胁迫响应和调节次生代谢产物等过程中起着重要作用。其中SbAbaIn、SbTDR和SbLHW.4g同时存在脱落酸、水杨酸、meJA反应、无氧诱导和光等相关元件,推测这3个基因可能更快响应多种环境胁迫,行使更多基因功能。
2.6 高粱与水稻、大豆、玉米和拟南芥MYC的系统发育分析将高粱、玉米、拟南芥、大豆和水稻MYC氨基酸序列进行聚类分析(图 6)。高粱28个MYC可分为8个亚族,高粱MYC在各亚族均有分布,集中分布在第Ⅳ亚族,且高粱与玉米MYC基因大多聚在同一分支下,表明高粱和玉米在进化过程中同源关系更为亲近。此外,SbR-S2与Araha.52607s0001互为直系同源基因,Araha.52607s000基因参与盐胁迫信号传导,推测SbR-S2可能发挥相似的作用。
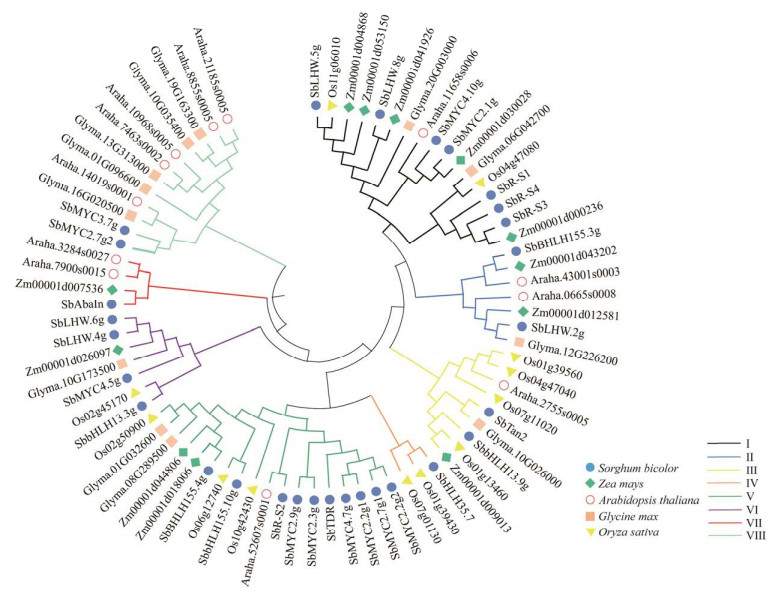
|
| 图 6 高粱与玉米、大豆、水稻和拟南芥MYC系统进化树分析 Fig. 6 Phylogenetic tree of MYCs of Sorghum bicolor, Zea may, Glycine max, Oryza sative and Arabidopsis thaliana. |
| |
遗传上使用非同义突变率(Ka)和同义突变率(Ks)的比值来判断此蛋白编码的基因是否有选择压力。如果Ka/Ks < 1,则认为有纯化选择作用。高粱MYC同源基因对分析进化选择压力见表 5,SbMYCs编码区基因的Ka/Ks均远小于1,说明SbMYCs蛋白编码区基因均受纯化选择作用,可以保证SbMYC蛋白功能的稳定性。
| Gene | Ka | Ks | Ka/Ks | Gene | Ka | Ks | Ka/Ks | |
| SbMYC2.1g | 0.028 953 | 0.213 777 | 0.135 435 | SbR-S4 | 0.115 402 | 0.316 814 | 0.364 259 | |
| SbLHW.2g | 0.105 211 | 0.257 736 | 0.408 212 | SbR-S3 | 0.121 883 | 0.368 833 | 0.330 454 | |
| SbTan2/GLABRA3 | 0.129 727 | 0.306 220 | 0.423 640 | SbR-S2 | 0.129 164 | 0.654 619 | 0.197 312 | |
| SbMYC2.2g2 | 0.036 775 | 0.104 750 | 0.351 075 | SbR-S1 | 0.140 073 | 0.617 129 | 0.226 976 | |
| SbMYC2.2g1 | 0.099 818 | 0.391 013 | 0.255 280 | SbLHW.6g | 0.043 988 | 0.174 795 | 0.251 657 | |
| SbbHLH13.3g | 0.031 224 | 0.385 553 | 0.080 985 | SbbHLH35.7g | 0.033 849 | 0.075 631 | 0.374 398 | |
| SbMYC2.3g | 0.325 899 | 0.574 851 | 0.566 928 | SbMYC2.7g1 | 0.103 717 | 0.277 024 | 0.742 392 | |
| SbAbaIn | 0.058 421 | 0.195 530 | 0.298 783 | SbMYC4.7g | 0.215 799 | 0.290 680 | 0.321 465 | |
| SbbHLH155.3g | 0.045 718 | 0.146 283 | 0.312 533 | SbMYC2.7g2 | 0.060 620 | 0.188 575 | 0.613 654 | |
| SbTDR | 0.024 558 | 0.115 933 | 0.211 827 | SbMYC3.7g | 0.072 284 | 0.117 793 | 0.278 681 | |
| SbbHLH155.4g | 0.103 001 | 0.196 459 | 0.524 288 | SbLHW.8g | 0.034 145 | 0.122 523 | 0.608 137 | |
| SbLHW.4g | 0.056 541 | 0.143 265 | 0.394 662 | SbMYC2.9g | 0.118 961 | 0.195 615 | 0.310 306 | |
| SbLHW.5g | 0.052 835 | 0.178 414 | 0.296 139 | SbbHLH13.9g | 0.091 333 | 0.294 332 | 0.246 551 | |
| SbMYC4.5g | 0.338 765 | 0.738 211 | 0.458 900 | SbbHLH155.10g | 0.056 885 | 0.230 724 | 0.321 465 |
利用高粱表达数据构建表达量热图(图 7),SbbHLH35.7g在生育期根、茎、叶和穗组织表达水平很高,在幼龄叶片中表达水平最高。SbLHW.2g在根组织表达水平比较高,SbLHW.8g在穗部表达水平较高,SbMYC2.1g、SbbHLH13.3g、SbLHW.6g和SbLHW.8g在种子内表达水平比较高。有些基因在整个高粱所有部位表达较低,如SbTan2、SbMYC4.5g、SbR-S4和SbR-S3等。SbTan2/GLABRA3是报道的控制籽粒单宁合成基因,SbR-S4、SbR-S1是报道的控制植株花青素合成基因,不表达很可能是所用材料TX623B发生了突变。

|
| 图 7 在高粱不同发育时期器官SbMYCs的表达 Fig. 7 Expression levels of SbMYCs in various tissues at different stages. |
| |
对高粱品系R111和SSA1不同发育阶段籽粒SbMYCs表达分析(图 8),发现发育早期籽粒中SbAbaIn表达最强,随籽粒发育降低。SbbHLH13.3g表达水平也较高。SbLHW.8g也有较高的相对表达量,20 d较10 d增加2倍。SbbHLH155.3g、SbTan2的表达水平也随籽粒发育表达增强。10 d籽粒SbTan2的表达水平R111明显高于SSA1。
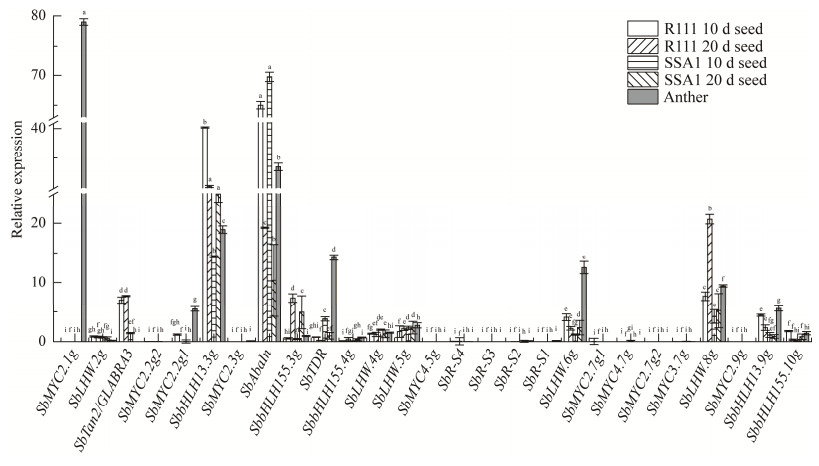
|
| 图 8 花粉和种子发育过程SbMYCs表达水平 Fig. 8 Expression levels of SbMYCs in pollen and various development stages of seeds. Different lowercase letters represent significant differences. 不同小写字母代表差异显著 |
| |
SbMYCs在成熟花粉中的表达分析显示,在花粉中显著表达的是SbMYC2.1g、SbAbaIn、SbbHLH13.3g、SbTDR、SbLHW.6g、SbLHW.8g和SbMYC2.2g1,SbbHLH13.9g次之。其他基因表达水平很低或不表达。
进一步测定了SbMYCs在抗蚜品系2457B、感蚜品系5-27sugB的5叶期叶片中表达水平,图 9A结果显示SbMYC2.1g和SbAbaIn表达水平较高。在抗、感蚜品系中均受到诱导表达的是SbAbaIn、SbLHW.4g和SbLHW.2g。仅在抗蚜品系显著诱导的有SbMYC2.1g和SbLHW.6g,在感蚜品系显著诱导的还有SbbHLH155.4g。SbTan2、SbR-S1和SbR-S2在叶片中几乎不表达。
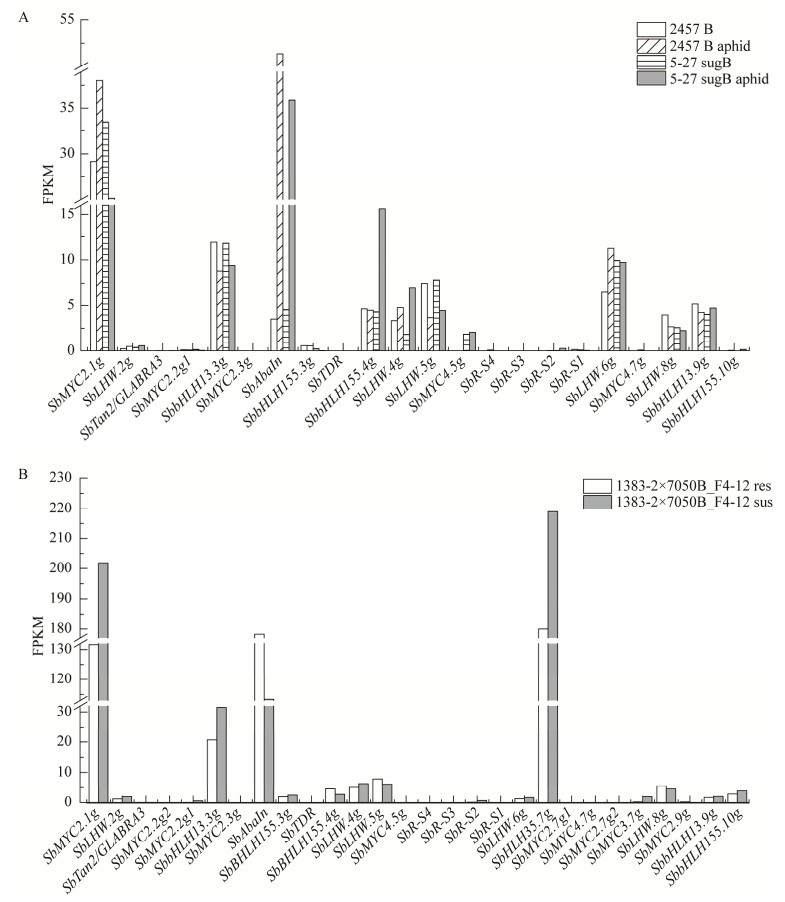
|
| 图 9 高粱蚜和丝轴黑粉菌侵染对高粱组织中SbMYCs表达的影响 Fig. 9 Effect of aphid and head smut infection on SbMYCs gene expression in sorghum tissues. A: Aphid infestation induced the expression of SbMYCs in sorghum leaves. B: Expression of SbMYCs in sorghum floral tissues infected by Sporisorium reilianum. A:高粱蚜侵染叶片. B:丝轴黑粉菌侵染穗部 |
| |
测定SbMYCs在抗、感丝黑穗病品系穗组织中的表达,图 9B结果显示SbbHLH35.7g在穗组织表达水平最高,其次是SbMYC2.1g和SbAbaIn。受侵染后SbbHLH35.7g表达水平受到显著诱导,SbMYC2.1g和SbHLH13.3g也得到诱导,在抗病品系表达明显较高的是SbAbaIn。
2.10 高粱MYC基因DNA自然等位变异通过Illumina全基因组重测序鉴定了所用种质的SNP和INDEL变异,见表 6。SbTan2在低单宁品系显示非同义SNP或INDEL阅读框改变。如在品系44B中,SbTan2在SBI-02的7 994 472处发生非同义突变,氨基酸由组氨酸突变为谷氨酰胺。SbTan2在SBI-02的7 994 628处发生移码突变,由G突变为GCCCTC。
| Gene | Reference | 2457B | 5-27A3R | R111 | 44B | Effect (other mutated line) |
| SbTan2/GLABRA3 | SBI-02: 7 994 472 T | T | T | T | G | Non-syn |
| SbTan2/GLABRA3 | SBI-02: 7 987 112 CCAA | CCAA | CCAA | CCAA | N | Codon deletion (V4A2) |
| SbTan2/GLABRA3 | SBI-02: 7 994 628 G | G | G | G | GCCCTC | Frame shift |
| SbMYC2.2g2 | SBI-02: 65 112 543 C to T | C | C | N | C | Non-syn (3030, C1-1) |
| SbbHLH155.3g | SBI-03: 72 859 488 ATT | AT | N | ATT | N | Upstream deletion |
| SbAbaIn | SBI-03: 60 870 863 G | G | R | A | N | Non-syn (1383-2) |
| SbR-S2 | SBI-06: 53 992 608G | T | T | G | G | Non-syn |
| SbR-S2 | SBI-06: 53 992 631C | G | G | C | C | Non-syn |
| SbR-S2 | SBI-06: 53 991 888 G | GTCC | GTCC | G | G | Codon insertion |
| SbMYC2.1g | SBI-01: 49 162 013 T | TCTGGTG | TCTGGTG | TCTGGTG | TCTGGTG | Codon insertion |
| SbTDR | SBI-04: 1 389 907 A | A | N | A | T | Non-syn |
| SbTDR | SBI-04: 1 389 989 G | GGGCGCCGCCGCC | G | N | G | Codon insertion |
| SbbHLH35.7g | SBI-07: 5 264 926 C | T | T | T | C | Non-syn |
| SbbHLH35.7g | SBI-07: 5 264446 CTCG | C | C | C | CTCG | Codon deletion |
为进一步挖掘SbTan2基因的自然变异,对该基因进行单倍型分析(图 10A),发现有6个SNP变异位点和7个INDEL变异位点,共10种单倍型。Hap1为主要单倍型,出现频次最多,共15次;Hap5出现2次。其余单倍型出现频次为1。在低单宁品系44B中发现4个INDEL变异、3个CODON_DELETION和1个FRAME_SHIFT,分别由编码核苷酸删除和插入引起的。
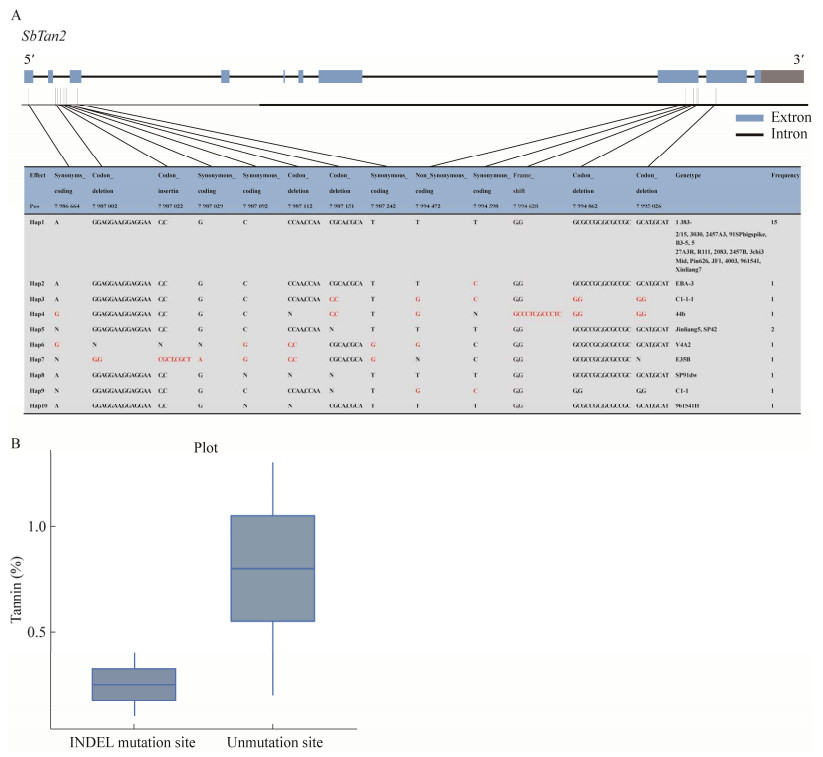
|
| 图 10 高粱SbTan2单倍型分析 Fig. 10 Haploid analysis of sorghum SbTan2. A: 25 haplotypes of SbTan2 gene in sorghum materials. B. Distribution of tannin content. A:25份高粱品系SbTan2基因单倍型. B:单宁含量分布 |
| |
进一步分析这4个变异与单宁含量的相关性(图 10B),发现SBI-02上7 987 151、7 994 628、7 994 862、7 995 026 bp位点处的INDEL变异,分别由CGGA突变为C,G移码突变为GCCCTC,GCGCCGC突变为G,由GCAT突变为G。且在25个品系中,高粱品系44B发生INDEL变异,系推测高粱品系44B的低单宁含量由于SbTan2的INDEL变异导致转录提前终止引起的。
2.12 SbMYC互作蛋白预测用STRING预测SbAbaIn和SbbHLH35.7g的互作蛋白(图 11),结果表明2种基因的互作蛋白差异较大,受蚜虫诱导表达的SbAbaIn与如下蛋白互作(表 7):4个Jasmonate ZIM domains (JAZs)结构域蛋白(TIFY),分别由SORBI_3006G056400、SORBI_3003G410300、SORBI_3002G036150和SORBI_3002G036100编码;3个锌指-FLZ结构域蛋白(FCS-like zinc finger 1-related, FLS)由SORBI_3006G194300、SORBI_3006G194100和SORBI_3004G278300编码,其中SORBI_3006G194300与红细胞转录因子GATA-1亚基A相似;还有由SORBI_3010G206400编码的转录因子Ets1、晚胚发生富集蛋白(late embryogenesis abundant protein, LEA-2)和ROH1-like。说明SbAbaIn通过茉莉酸(jasmonic acid, JA)途径提高高粱抵抗蚜虫侵染。
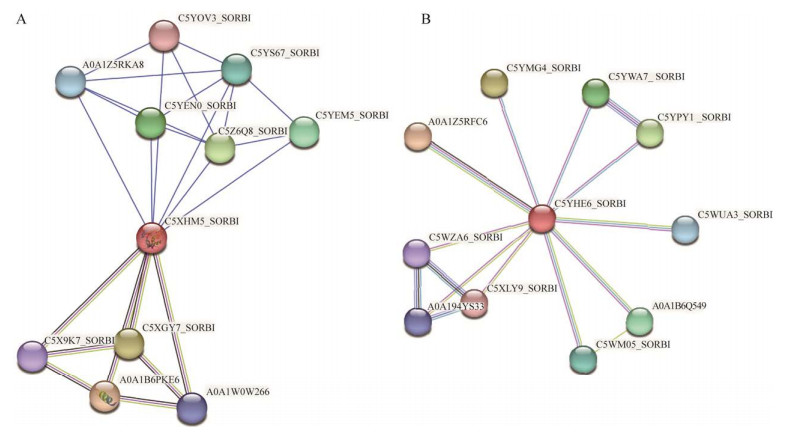
|
| 图 11 STRING预测SbAbaIn (A)和SbbHLH35.7g (B)蛋白互作网络 Fig. 11 STRING prediction of protein interaction network of SbAbaIn (A) and SbbHLH35.7g (B). |
| |
| 5XHM5, SbAbaIn | C5YHE6, SbbHLH35.7g | |||||
| Predicted functional partners | Annotation | Score | Predicted functional partners | Annotation | Score | |
| A0A1B6PKE6 SORBI_3006G056400 | Jasmonate ZIM domain-containing protein, TIFY domain-containing protein | 0.530 | A0A1Z5RFC6 SORBI_3006G179400 | Multidrug efflux transporter (MDR), cholesterol transporter 1, SSD domain-containing protein | 0.977 | |
| C5XGY7 SORBI_3003G410300 | Jasmonate ZIM domain-containing protein, TIFY domain-containing protein | 0.530 | C5YMG4 SORBI_3007G022000 |
Importin N-terminal domain-containing protein | 0.761 | |
| C5Z6Q8 SORBI_3010G206400 | Transcription factor Ets-1, SAM domain-containing protein | 0.512 | C5YPY1 SORBI_3008G135200 | Importin N-terminal domain-containing protein | 0.761 | |
| C5YEN0 SORBI_3006G194300 |
Erythroid transcription factor GATA-1 subunit A, Zf-FLZ-type domain-containing protein | 0504 | C5YWA7 SORBI_3009G102900 | Importin N-terminal domain-containing protein | 0.761 | |
| C5YEM5 SORBI_3006G194100 |
FCS-like zinc finger 1-related, Zf-FLZ-type domain-containing protein | 0.494 | A0A1B6Q549 SORBI_3003G241100 | Endopeptidase S2P, peptidase_M50 domain-containing protein | 0.705 | |
| C5YS67 SORBI_3008G042900 |
Late embryogenesis abundant protein LEA-2, LEA_2 domain-containing protein | 0.484 | C5WM05 SORBI_3001G104900 | Zinc metallopeptidase egy3, chloroplastic-related, peptidase M50 domain-containing protein | 0.705 | |
| A0A1Z5RKA8 SORBI_3004G006500 |
Tranmembrane helix, ROH1-like, BPS1 | 0.483 | C5WUA3 SORBI_3001G183000 | Metalloendopeptidase, PDZ domain-containing protein | 0.705 | |
| A0A1W0W266 SORBI_3002G036150 |
Jasmonate ZIM domain-containing protein, TIFY domain-containing protein | 0.480 | A0A194YS33 SORBI_3004G216200 | Aurora kinase, protein kinase domain-containing protein | 0.602 | |
| C5X9K7 SORBI_3002G036100 |
Jasmonate ZIM domain-containing protein, TIFY domain-containing protein | 0.480 | C5WZA6 SORBI_3001G078600 | Aurora kinase, protein kinase domain-containing protein | 0.602 | |
| C5Y0V3 SORBI_3004G278300 | FCS-like zinc finger 1-related, Zf-FLZ-type domain-containing protein | 0.476 | C5XLY9 SORBI_3003G034600 | Aurora kinase, protein kinase domain-containing protein | 0.602 | |
受丝轴黑粉菌诱导的SbbHLH35.7g与如下蛋白互作:多药剂外泵蛋白(multidrug efflux transporter, MDR)由SORBI_3006G179400编码;3个核转运蛋白importin由SORBI_3007G022000、SORBI_3008G135200和SORBI_3009G102900编码;3个aurora激酶由SORBI_3004G216200、SORBI_3001G078600和SORBI_3003G034600编码;3个多肽酶由SORBI_3001G183000、SORBI_3003G241100和SORBI_3001G104900编码。推测SbbHLH35.7g通过毒素外运、降解等途径提高高粱防御能力。
3 讨论与结论高粱是典型的杂粮作物,但易遭受蚜虫、丝黑穗病等生物胁迫,遭受胁迫后的植株长势变弱且生长缓慢,穗发育受阻,严重降低生物量和产量。已有研究表明,MYC基因在植物抗性和次生代谢产物积累方面起重要作用,分析高粱MYC的功能对提高抗性、产量与品质有重大意义。
已有研究表明,MYC转录因子调节植物抗性与生长发育的过程受JA信号调控,MYC2也是脱落酸ABA信号转导的正调节因子,已被证明与ABA介导的发芽抑制相关[41]。大多高粱MYC基因家族启动子区富含4种顺式元件ABRE、CGTCA-motif、G-Box和TGACG-motif。SbMYC2.1g和SbAbaIn在5叶期高粱叶片中显著表达,后者显著受高粱蚜诱导。SbbHLH35.7g在穗组织表达水平最高,受丝轴黑粉菌侵染后显著上调,推测是MYC基因参与了JA信号转导,茉莉酸通路的相应顺式元件可使其响应逆境胁迫,从而调节生长发育。SbMYCs启动子区包含茉莉酸甲酯、脱落酸、水杨酸、光和干旱诱导相关等顺式作用元件等,表明此基因家族成员能更快地响应光反应和逆境胁迫。
热图分析表明,SbbHLH35.7g在生育期多数组织中表达水平比较高,在叶片和茎中表达水平达到最高,说明这个部位代谢活动强,具有重要功能。SbR-S2在SBI-06上存在2个突变位点,SBI-06上有叶鞘色基因Rs和成株颜色Q[18],SbR-S1和SbR-S3 (Sobic.006G175700、Sobic.006G175200)是已鉴定的叶片花青素合成调控基因,本研究通过转录组数据分析发现其有多达25个转录本,编码MYB的Sobic.005G014100也影响花青素合成。Chopra等[24]发现Sobic.006G025040属于bHLH转录因子,与叶片的紫色色素沉着有关,也在高粱抗冷性上发挥关键作用。MYC参与受伤反应,可提高抗旱、抗病虫能力[42]。
高粱籽粒发育过程中SbMYCS表达水平呈升高或降低2种模式。SbABAIn在籽粒中表达最强,随籽粒发育降低;而SbbHLH13.3g、SbLHW.8g、SbbHLH155.3g和SbTan2的表达水平随籽粒发育表达增强;SbTan2的表达水平因品种的单宁含量而异。Wu等[22]发现SbTan2/ Sobic.002G076600调控单宁合成,指出Tx430的SbTan2 INDEL变异导致转录提前终止。这在本研究得到证实,同时发现有些无、低单宁基因型存在非同义SNP。Ayalew等[43]鉴定到与籽粒蛋白含量关联的bHLH基因,包括Sobic.001G416500、Sobic.001G349700和Sobic.001G350300。Morris等[44]鉴定到籽粒积累相关的多酚合成的控制位点,包括黄酮代谢酶F3′H基因Sobic.004G200800、Sobic.004G200900和Sobic.004G201100。Pinheiro等[45]指出干旱可增加无单宁高粱的黄酮和3-脱氧花青素,提高了其籽粒功能特征。通过诱变育种,已经获得富含3-脱氧花青素的红叶突变体RG,但并未报道受突变的基因[46]。Mizuno等[47]指出切割诱导植株褐色形成机制,黄酮合成酶Ⅱ (FNSII)的表达导致芹菜素(apigenin)合成,表达FNSII和黄酮类3′-羟化酶(flavonoid 3′-hydroxylase, F3′H)导致apigenin和木犀草素(luteolin)合成,表达黄烷酮4-还原酶和F3′H导致合成3-脱氧花青素。这些为解析高粱籽粒和植株颜色提供了很好的范例。高粱生产主要依赖于雄性不育基础上的杂种优势,但对花粉发育、雄性不育形成的分子机制了解较少[48-49]。本研究鉴定到SbTDR (ZmMS40)与SbEAR等水稻、玉米花粉发育同源基因[50-51],发现花粉中表达最强的是SbMYC2.1g,其次是SbAbaIn、SbbHLH13.3g和SbTDR,同时找到了这些基因的SNP或INDEL标记,有助于高粱育性研究。
本研究从高粱基因组鉴定到28个MYC基因,其表达存在时空差异。SbMYCs受蚜虫和丝黑穗诱导表达,在抗、感蚜品系显著诱导的是SbAbaIn、SbLHW.4g和SbLHW.2g。受丝轴黑粉菌侵染后SbbHLH35.7g、SbMYC2.1g和SbHLH13.3g显著表达,且侵染后在花粉中表达水平较强的是SbMYC2.1g、SbAbaIn、SbbHLH13.3g和SbTDR等。SbTan2、SbR-S4等的SNP、Indel变异,为高粱性状形成的分子机制奠定了基础。
致谢: 山西农业大学(山西省农业科学院)高粱研究所张福耀研究员和柳青山研究员分别提供了SSA1和1383-2原始种子,特此致谢。
| [1] |
李顺国, 刘猛, 刘斐, 邹剑秋, 陆晓春, 刁现民. 中国高粱产业和种业发展现状与未来展望[J]. 中国农业科学, 2021, 54(3): 471-482. LI SG, LIU M, LIU F, ZOU JQ, LU XC, DIAO XM. Current status and future prospective of sorghum production and seed industry in China[J]. Scientia Agricultura Sinica, 2021, 54(3): 471-482 (in Chinese). |
| [2] |
谢鹏飞, 朱蕾, 冯玲, 吴进才, 刘景澜. 转录因子MYC2介导植物抗生物胁迫的研究进展[J]. 应用昆虫学报, 2020, 57(4): 781-787. XIE PF, ZHU L, FENG L, WU JC, LIU JL. Research progress in transcription factor MYC2 mediating plant resistance to biological stress[J]. Chinese Journal of Applied Entomology, 2020, 57(4): 781-787 (in Chinese). |
| [3] |
GAO CH, QI SH, LIU KG, LI D, JIN CY, LI ZW, HUANG GQ, HAI JB, ZHANG M, CHEN MX. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis[J]. Plant Physiology and Biochemistry: PPB, 2016, 108: 63-70. DOI:10.1016/j.plaphy.2016.07.004
|
| [4] |
PIRES N, DOLAN L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Molecular Biology and Evolution, 2010, 27(4): 862-874. DOI:10.1093/molbev/msp288
|
| [5] |
LEDENT V, VERVOORT M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis[J]. Genome Research, 2001, 11(5): 754-770. DOI:10.1101/gr.177001
|
| [6] |
STRYGINA KV, KHLESTKINA EK. Myc-like transcriptional factors in wheat: structural and functional organization of the subfamily I members[J]. BMC Plant Biology, 2019, 19(1): 59-69. DOI:10.1186/s12870-019-1656-7
|
| [7] |
ZHANG M, JIN XF, CHEN Y, WEI M, LIAO WF, ZHAO SY, FU CH, YU LJ. TcMYC2a, a basic helix-loop-helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis[J]. Frontiers in Plant Science, 2018, 9: 863. DOI:10.3389/fpls.2018.00863
|
| [8] |
LUDWIG SR, HABERA LF, DELLAPORTA SL, WESSLER SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region[J]. Proceedings of the National Academy of Sciences of the United States of America, 1989, 86(18): 7092-7096.
|
| [9] |
HICHRI I, HEPPEL SC, PILLET J, LÉON C, CZEMMEL S, DELROT S, LAUVERGEAT V, BOGS J. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine[J]. Molecular Plant, 2010, 3(3): 509-523. DOI:10.1093/mp/ssp118
|
| [10] |
SRIVASTAVA M, SRIVASTAVA AK, ROY D, MANSI MS, GOUGH C, BHAGAT PK, ZHANG CJ, SADANANDOM A. The conjugation of SUMO to the transcription factor MYC2 functions in blue light-mediated seedling development in Arabidopsis[J]. The Plant Cell, 2022, 34(8): 2892-2906. DOI:10.1093/plcell/koac142
|
| [11] |
YADAV V, MALLAPPA C, GANGAPPA SN, BHATIA S, CHATTOPADHYAY S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth[J]. The Plant Cell, 2005, 17(7): 1953-1966. DOI:10.1105/tpc.105.032060
|
| [12] |
VERMA D, JALMI SK, BHAGAT PK, VERMA N, SINHA AK. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis[J]. The FEBS Journal, 2020, 287(12): 2560-2576. DOI:10.1111/febs.15157
|
| [13] |
ABE H, YAMAGUCHI-SHINOZAKI K, URAO T, IWASAKI T, HOSOKAWA D, SHINOZAKI K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression[J]. The Plant Cell, 1997, 9(10): 1859-1868.
|
| [14] |
CHINI A, MONTE I, ZAMARREÑO AM, HAMBERG M, LASSUEUR S, REYMOND P, WEISS S, STINTZI A, SCHALLER A, PORZEL A, GARCÍA-MINA JM, SOLANO R. An OPR3-independent pathway uses 4, 5-didehydrojasmonate for jasmonate synthesis[J]. Nature Chemical Biology, 2018, 14(2): 171-178. DOI:10.1038/nchembio.2540
|
| [15] |
YU ZX, LI JX, YANG CQ, HU WL, WANG LJ, CHEN XY. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L.[J]. Molecular Plant, 2012, 5(2): 353-365. DOI:10.1093/mp/ssr087
|
| [16] |
YANG YF, ZHANG KK, YANG LY, LV X, WU Y, LIU HW, LU Q, CHEN DF, QIU DY. Identification and characterization of MYC transcription factors in Taxus sp.[J]. Gene, 2018, 675: 1-8. DOI:10.1016/j.gene.2018.06.065
|
| [17] |
ZHONG JY, JIN ZH, JIANG L, ZHANG LX, HU ZT, ZHANG YH, LIU YB, MA JB, HUANG Y. Structural basis of the bHLH domains of MyoD-E47 heterodimer[J]. Biochemical and Biophysical Research Communications, 2022, 621: 88-93. DOI:10.1016/j.bbrc.2022.06.071
|
| [18] |
MACE ES, JORDAN DR. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench)[J]. Theoretical and Applied Genetics, 2010, 121(7): 1339-1356. DOI:10.1007/s00122-010-1392-8
|
| [19] |
张春来, 李艳锋, 赵威军, 赵靓, 王晨, 梁笃, 周福平. 高粱品质性状改良的分子遗传学基础[J]. 植物生理学报, 2015(5): 610-616. ZHANG CL, LI YF, ZHAO WJ, ZHAO L, WANG C, LIANG D, ZHOU FP. Molecular genetic basis for biotechnological improvement of grain quality characteristics in sorghum[J]. China Industrial Economics, 2015(5): 610-616 (in Chinese). |
| [20] |
冯周, 曹宁, 丁延庆, 徐建霞, 高旭, 程斌, 邹桂花, 张立异. 利用超高密度Bin图谱定位高粱籽粒物理性状的QTL[J]. 植物遗传资源学报, 2022, 23(6): 1746-1755. FENG Z, CAO N, DING YQ, XU JX, GAO X, CHENG B, ZOU GH, ZHANG LY. Mapping QTL for sorghum physical properties using ultra-high-density Bin map[J]. Journal of Plant Genetic Resources, 2022, 23(6): 1746-1755 (in Chinese). |
| [21] |
XIE P, SHI JY, TANG SY, CHEN CX, KHAN A, ZHANG FX, XIONG Y, LI C, HE W, WANG GD, LEI FM, WU YR, XIE Q. Control of bird feeding behavior by tannin1 through modulating the biosynthesis of polyphenols and fatty acid-derived volatiles in sorghum[J]. Molecular Plant, 2019, 12(10): 1315-1324. DOI:10.1016/j.molp.2019.08.004
|
| [22] |
WUYY, GUO TT, MU Q, WANG JY, LI X, WU Y, TIAN B, WANG ML, BAI GH, PERUMAL R, TRICK HN, BEAN SR, DWEIKAT IM, TUINSTRA MR, MORRIS G, TESSO TT, YU JM, LI XR. Allelochemicals targeted to balance competing selections in African agroecosystems[J]. Nature Plants, 2019, 5(12): 1229-1236. DOI:10.1038/s41477-019-0563-0
|
| [23] |
LV Y, CHEN J, ZHU MJ, LIU YS, WU XY, XIAO X, YUYAMA N, LIU FX, JING HC, CAI HW. Wall-associated kinase-like gene RL1 contributes to red leaves in sorghum[J]. The Plant Journal: for Cell and Molecular Biology, 2022, 112(1): 135-150. DOI:10.1111/tpj.15936
|
| [24] |
CHOPRA R, BUROW G, BURKE JJ, GLADMAN N, XIN ZG. Genome-wide association analysis of seedling traits in diverse sorghum germplasm under thermal stress[J]. BMC Plant Biology, 2017, 17(1): 1-15. DOI:10.1186/s12870-016-0951-9
|
| [25] |
ELANGO D, XUE WY, CHOPRA S. Genome wide association mapping of epi-cuticular wax genes in Sorghum bicolor[J]. Physiology and Molecular Biology of Plants, 2020, 26(8): 1727-1737. DOI:10.1007/s12298-020-00848-5
|
| [26] |
FAN Y, YANG H, LAI DL, HE AL, XUE GX, FENG L, CHEN L, CHENG XB, RUAN JJ, YAN J, CHENG JP. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in sorghum (Sorghum bicolor (L.) Moench)[J]. BMC Genomics, 2021, 22(1): 415. DOI:10.1186/s12864-021-07652-9
|
| [27] |
SONG YS, LI SM, SUI Y, ZHENG HX, HAN GL, SUN X, YANG WJ, WANG HL, ZHUANG KY, KONG FY, MENG QW, SUI N. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum[J]. Theoretical and Applied Genetics, 2022, 135(1): 201-216. DOI:10.1007/s00122-021-03960-6
|
| [28] |
刘国庆, 杜瑞恒, 侯升林, 吕芃, 籍贵苏, 李素英. 高粱抗蚜研究进展[J]. 植物学报, 2012, 47(2): 171-187. LIU GQ, DU RH, HOU SL, LV F, JI GS, LI SY. Research progress on sorghum resistance to aphid[J]. Chinese Journal of Botany, 2012, 47(2): 171-187 (in Chinese). |
| [29] |
邹剑秋, 李玥莹, 朱凯, 王艳秋. 高粱丝轴黑粉菌3号生理小种抗性遗传研究及抗病基因分子标记[J]. 中国农业科学, 2010, 43(4): 713-720. ZOU JQ, LI YY, ZHU K, WANG YQ. Study on inheritance and molecular markers of sorghum resistance to head smut physiological race 3[J]. Scientia Agricultura Sinica, 2010, 43(4): 713-720 (in Chinese). DOI:10.3864/j.issn.0578-1752.2010.04.007 |
| [30] |
WANG FM, ZHAO SM, HAN YH, SHAO YT, DONG ZY, GAO Y, ZHANG KP, LIU X, LI DW, CHANG JH, WANG DW. Efficient and fine mapping of RMES1 conferring resistance to sorghum aphid Melanaphis sacchari[J]. Molecular Breeding, 2013, 31(4): 777-784. DOI:10.1007/s11032-012-9832-6
|
| [31] |
MCCORMICK RF, TRUONG SK, SREEDASYAM A, JENKINS J, SHU SQ, SIMS D, KENNEDY M, AMIREBRAHIMI M, WEERS BD, McKINLEY B, MATTISON A, MORISHIGE DT, GRIMWOOD J, SCHMUTZ J, MULLET JE. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization[J]. The Plant Journal: for Cell and Molecular Biology, 2018, 93(2): 338-354. DOI:10.1111/tpj.13781
|
| [32] |
张晓. 高粱单宁调控基因Tannin2互作蛋白的鉴定及分子标记开发[D]. 泰安: 山东农业大学硕士学业论文, 2023. ZHANG X. Identification of protein interacting with Tannin2 gene and development of related-molecular markers[D]. Taian: Master's thesis of Shandong Agricultural Aniversity, 2023 (in Chinese). |
| [33] |
CHEN CJ, CHEN H, ZHANG Y, THOMAS HR, FRANK MH, HE YH, XIA R. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202. DOI:10.1016/j.molp.2020.06.009
|
| [34] |
CHENNA R, SUGAWARA H, KOIKE T, LOPEZ R, GIBSON TJ, HIGGINS DG, THOMPSON JD. Multiple sequence alignment with the clustal series of programs[J]. Nucleic Acids Research, 2003, 31(13): 3497-3500. DOI:10.1093/nar/gkg500
|
| [35] |
TAMURA K, PETERSON D, PETERSON N, STECHER G, NEI M, KUMAR S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10): 2731-2739. DOI:10.1093/molbev/msr121
|
| [36] |
BAILEY TL, BODEN M, BUSKE FA, FRITH M, GRANT CE, CLEMENTI L, REN JY, LI WW, NOBLE WS. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Research, 2009, 37(web server issue): W202-W208.
|
| [37] |
ARTIMO P, JONNALAGEDDA M, ARNOLD K, BARATIN D, CSARDI G, de CASTRO E, DUVAUD S, FLEGEL V, FORTIER A, GASTEIGER E, GROSDIDIER A, HERNANDEZ C, IOANNIDIS V, KUZNETSOV D, LIECHTI R, MORETTI S, MOSTAGUIR K, REDASCHI N, ROSSIER G, XENARIOS I, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Research, 2012, 40(web server issue): W597-W603.
|
| [38] |
GEOURJON C, DELÉAGE G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Computer Applications in the Biosciences: CABIOS, 1995, 11(6): 681-684.
|
| [39] |
赵珊珊, 郭志强, 朱立勋, 范佳利, 杨博慧, 柴文婷, 孙慧琼, 冯凡, 粱月秀, 邹春雷, 姜晓东, 赵威军, 吕晋慧, 张春来. 高粱高亲和硝酸盐转运蛋白NRT2/3基因家族鉴定、表达与DNA变异分析[J]. 生物工程学报, 2023, 39(7): 2743-2761. ZHAO SS, GUO ZQ, ZHU LX, FAN JL, YANG BH, CHAI WT, SUN HQ, FENG F, LIANG YX, ZOU CL, JIANG XD, ZHAO WJ, LÜ JH, ZHANG CL. Identification, expression and DNA variation analysis of high affinity nitrate transporter NRT2/3 gene family in Sorghum bicolor[J]. Chinese Journal of Biotechnology, 2023, 39(7): 2743-2761 (in Chinese). |
| [40] |
范佳利, 朱立勋, 郭志强, 尹梦娇, 杨博慧, 柴文婷, 赵珊珊, 孙慧琼, 李莎莎, 丁鹏程, 王爱萍, 姜晓东, 贾举庆, 杨珍平, 吕晋慧, 高志强, 张春来. 普通小麦JAZ基因家族鉴定及其响应冻害时基因差异表达和DNA变异分析[J]. 植物生理学报, 2022, 58(10): 1873-1889. FAN JL, ZHU LX, GUO ZQ, YIN MJ, YANG BH, CHAI WT, ZHAO SS, SUN HQ, LI SS, DING PC, WANG AP, JIANG XD, JIA JQ, YANG ZP, LÜ JH, GAO ZQ, ZHANG CL. Identification of JAZ gene family and analysis on their differential expression under freezing stress and DNA variation in common wheat[J]. Plant Physiology Journal, 2022, 58(10): 1873-1889 (in Chinese). |
| [41] |
ALEMAN F, YAZAKI J, LEE M, TAKAHASHI Y, KIM AY, LIZX, KINOSHITA T, ECKER JR, SCHROEDER JI. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling[J]. Scientific Reports, 2016, 6: 28941. DOI:10.1038/srep28941
|
| [42] |
TREUTTER D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis[J]. Plant Biology, 2005, 7(6): 581-591. DOI:10.1055/s-2005-873009
|
| [43] |
AYALEW H, PEIRIS S, CHILUWAL A, KUMAR R, TIWARI M, OSTMEYER T, BEAN S, JAGADISH SVK. Stable sorghum grain quality QTL were identified using SC35×RTx430 mapping population[J]. The Plant Genome, 2022, 15(3): e20227. DOI:10.1002/tpg2.20227
|
| [44] |
MORRIS GP, RHODES DH, BRENTON Z, RAMU P, THAYIL VM, DESHPANDE S, ACHARYA C, MITCHELL SE, BUCKLER ES, YU JM, KRESOVICH S. Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits[J]. G3 (Bethesda, Md), 2013, 3(11): 2085-2094. DOI:10.1534/g3.113.008417
|
| [45] |
PINHEIRO SS, de MORAIS CL, ANUNCIAÇÃO PC, de MENEZES CB, VIEIRA QVA, MARTINO HSD, DELLA LCM, PINHEIRO SAHM. Water stress increased the flavonoid content in tannin-free sorghum grains[J]. Journal of Food Composition and Analysis, 2021, 100: 103892. DOI:10.1016/j.jfca.2021.103892
|
| [46] |
PETTI C, KUSHWAHA R, TATENO M, ELIZABETH HARMAN-WARE A, CROCKER M, AWIKA J, DeBOLT S. Mutagenesis breeding for increased 3-deoxyanthocyanidin accumulation in leaves of Sorghum bicolor (L.) Moench: a source of natural food pigment[J]. Journal of Agricultural and Food Chemistry, 2014, 62(6): 1227-1232. DOI:10.1021/jf405324j
|
| [47] |
MIZUNO H, YAZAWA T, KASUGA S, SAWADA Y, KANAMORI H, OGO Y, HIRAIMY, MATSUMOTO T, KAWAHIGASHI H. Expression of Flavone synthase Ⅱ and Flavonoid 3'-hydroxylase is associated with color variation in Tan-colored injured leaves of sorghum[J]. Frontiers in Plant Science, 2016, 7: 1718.
|
| [48] |
DHAKA N, SHARMA S, VASHISHT I, KANDPAL M, SHARMA MK, SHARMA R. Small RNA profiling from meiotic and post-meiotic anthers reveals prospective miRNA-target modules for engineering male fertility in sorghum[J]. Genomics, 2020, 112(2): 1598-1610. DOI:10.1016/j.ygeno.2019.09.009
|
| [49] |
KANTE M, RATTUNDE HFW, NÉBIÉ B, WELTZIEN E, HAUSSMANN BIG, LEISER WL. QTL mapping and validation of fertility restoration in west African sorghum A1 cytoplasm and identification of a potential causative mutation for Rf2[J]. Theoretical and Applied Genetics, 2018, 131(11): 2397-2412. DOI:10.1007/s00122-018-3161-z
|
| [50] |
KIYOSAWA A, YONEMARUJI, MIZUNO H, KANAMORI H, WU JZ, KAWAHIGASHI H, GOTO K. Fine mapping of Rf5 region for a sorghum fertility restorer gene and microsynteny analysis across grass species[J]. Breeding Science, 2022, 72(2): 141-149. DOI:10.1270/jsbbs.21057
|
| [51] |
KO SS, LI MJ, KU MSB, HO YC, LIN YJ, CHUANG MH, HSING HX, LIEN YC, YANG HT, CHANG HC, CHAN MT. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice[J]. The Plant Cell, 2014, 26(6): 2486-2504. DOI:10.1105/tpc.114.126292
|
 2024, Vol. 40
2024, Vol. 40