中国科学院微生物研究所、中国微生物学会主办
文章信息
- Amna Bibi, 苏立秋, 戴宗杰, 王钦宏
- Amna Bibi, SU Liqiu, DAI Zongjie, WANG Qinhong
- 萜类化合物微生物合成中酶工程的研究进展与展望
- Enzyme engineering in microbial biosynthesis of terpenoids: progress and perspectives
- 生物工程学报, 2024, 40(8): 2473-2488
- Chinese Journal of Biotechnology, 2024, 40(8): 2473-2488
- 10.13345/j.cjb.240165
-
文章历史
- Received: February 28, 2024
- Accepted: May 8, 2024
2. 国家合成生物技术创新中心, 天津 300308;
3. 中国科学院大学, 北京 100049
2. National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China;
3. University of Chinese Academy of Science, Beijing 100049, China
Terpenoids are highly diverse natural compounds, with over 80 000 distinct structures. They can be found in a broad range of organisms, including plants, fungi, bacteria, marine animals, insects, and archaea[1]. Although certain terpenes are largely responsible for protective toxins, others also have pleasant aromas. As a result, they have attracted a lot of interests in the food, drug, and cosmetic industries[2]. Terpenoids can be classified into distinct groups based on the number of isoprene (C5) units they possess. These groups include hemiterpenoids (C5), monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), sesterterpenoids (C25), triterpenoids (C30), tetraterpenoids (C40), and polyterpenoids (C > 40). Terpenoids can also be synthesized chemically, but these processes are energy intensive, costly and produce a lot of organic waste that is also unfriendly to the environment. The production of terpenoids using microbial hosts is considered as a safe, cost-competitive and scalable approach[3]. Biosynthesis of terpenoids employs dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) as building blocks, which are five-carbon isomers. These building blocks are produced through either the mevalonate (MVA) pathway or the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway in biological systems[4]. Prenyl diphosphate synthases then condense these building blocks to produce prenyl chains of varying sizes, with geranyl diphosphate (GPP), farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP), serving as the substrates of terpene synthases (Figure 1).

|
| 图 1 萜类微生物合成途径 Fig. 1 Pathways for microbial synthesis of terpenoids. Highlighting key steps of upstream precursor supply by mevalonate (MVA) and 2-C-methyl-d-erythritol-4-phosphate (MEP) pathways, downstream terpene synthesis by exogenous terpenoid synthetic pathway, and engineered enzymes used for monoterpenes, sesquiterpenes, and diterpenes production discussed in this review are shown in red. G3P: Glyceraldehyde-3-phosphate; DXP: 1-deoxy-d-xylulose 5-phosphate; CDP-ME: 4-diphosphocytidyl-2C-methyl-d-erythritol; CDP-MEP: 4-diphosphocytidyl-2C-methyl-d-erythritol phosphate; MEP-CPP: 2C-methyl-d-erythritol 2, 4-cyclodiphosphate; HMBPP: 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; IPP: Isopentenyl pyrophosphate; DMAPP: Dimethylallyl pyrophosphate; GPP: Geranyl disphosphate; FPP: Farnesyl diphosphate; GGPP: Geranylgeranyl diphosphate. |
| |
Many efforts have been made in recent years to improve the production of terpenoid in microbes, such as Saccharomyces cerevisiae and Escherichia coli[5-6]. However, poor productivity (low titer, low yield, and low efficiency) and lack of stability of microorganisms during fermentation, is currently posing a challenge to establish strains for commercial production of terpenoids[7]. Competition between the heterologous terpenoid biosynthetic route and inherent metabolic processes of the host strain is a significant barrier to high-level terpenoid synthesis. Accumulation of hydrophobic terpenoids may contribute to the metabolic stress and damage the cells, causing cytotoxicity. A further major concern is the ineffective catalytic capability of terpenoid-related enzymes, which reduces the productivity of target products and necessitates an expensive purification procedure[8]. Therefore, in the context of heterologous microbial system, enzyme modification is crucial for refining enzyme activity beyond natural capabilities. In this study, we will shed some lights on several terpenoids biosynthesis related enzymes and highlight the significance of enzyme engineering in the yield of terpenoids via microbial systems.
1 Enzyme engineering strategies for terpenoids productionEnzyme engineering plays a crucial role in enhancing enzyme activity, stability, and broadening the availability of terpenoid biosynthetic products[9]. Among the key methods that have emerged are directed evolution, rational design, and semi-rational modification. Directed evolution, agnostic to enzyme structure and catalytic mechanisms, utilizes mutagenesis techniques such as error-prone PCR, DNA shuffling, and site-directed mutagenesis to create a diverse mutant library. Subsequently, selection pressures are applied to identify mutants with desired characteristics. Its advantage lies in its ability to transcend limitations in protein structure-function comprehension. However, it can be resource-intensive and requires efficient high-throughput screening methodologies[10-11]. In contrast, rational design heavily relies on knowledge of protein sequences, structures, and catalytic mechanisms. It involves meticulous modifications, leveraging computational tools such as molecular docking, dynamics simulations, and quantum mechanics. Rational design's primary benefit is its efficiency in narrowing down the mutant pool and targeting specific objectives. Nevertheless, its success is often constrained by incomplete knowledge of protein structures and the complexities of predicting mutational interactions[12]. Semi-rational design serves as a bridge between directed evolution and rational design. It integrates rational design principles within a directed evolution framework, confining random mutations to specific amino acid residues. This approach ensures a smaller mutant library, thereby increasing the likelihood of obtaining favorable mutants. However, it still requires a certain level of understanding of protein function and structure[10] (Figure 2). Each protein engineering strategy has unique strengths and weaknesses. The choice of strategy hinges on the specific objectives, the extant knowledge about the protein, and the available resources.
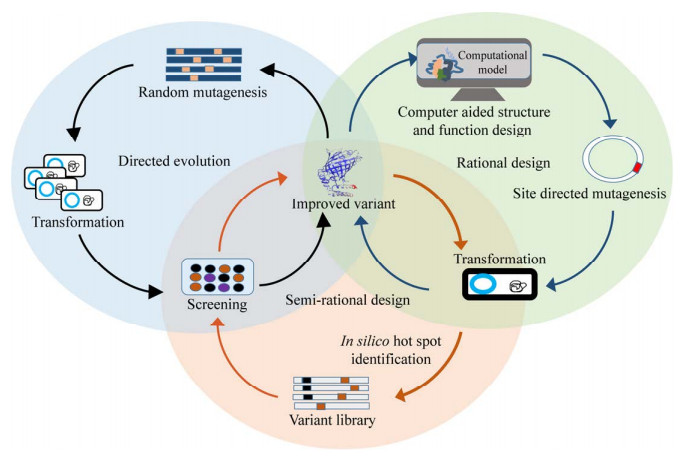
|
| 图 2 蛋白质工程策略 Fig. 2 General strategies for protein engineering. Protein engineering primarily relies on directed evolution, rational engineering, and semi-rational design to effectively construct the desired enzyme variants. |
| |
In addition to the main strategies stated above, protein engineering involves various other strategies to modify and optimize proteins for specific purposes, including protein fusion and chemical modification. Protein fusion combines different proteins to create hybrids with new or improved properties. Chemical modification alters amino acid residues to achieve the desired changes[13]. These enzyme engineering techniques have been particularly beneficial in promoting the microbial biosynthesis of terpenoids. They enable the optimization of enzyme stability, substrate specificity, product specificity, catalytic efficiency, and other desired characteristics. By employing these strategies, researchers can enhance the production and yield of terpenoids.
1.1 Enhancing catalytic efficiency 1.1.1 Modifications of active centerFor the overall catalytic outcome, the amino acid residues of active center, are of great value. They play a vital role in shaping the modulation and variations of active sites as well. One crucial enzyme in the terpenoids biosynthesis pathway is mevalonate kinase (MK). To optimize MK activity, a study employed a directed evolution strategy in E. coli, and three specific mutations (V13D, S148I, and V301E) which are located in the active center were identified[14]. Compared to the wild-type MK, the purified mutant MK exhibited a decrease of 74% in the value of Km and an improvement of 26% in the value of kcat, indicating improved substrate affinity and enhanced catalytic efficiency of the mutant enzyme[14]. These improvements led to a remarkable 2.4-fold increase in lycopene production when using the mutant MK compared to the wild-type MK (Figure 3A)[14].

|
| 图 3 酶工程改造策略 Fig. 3 Various enzyme engineering strategies employed to enhance catalytic activity (A), stability (B), substrate and product specificity (C and D), enzyme fusion (E) and colocalization (F). CrtW: 4, 4′ β-carotene oxygenase; CrtZ: 3, 3′ β-carotene hydroxylase. 提高催化活性(A), 提高酶稳定性(B), 提高底物(C)及产物(D)特异性, 蛋白连接器促进酶融合(E), 蛋白脚手架促进酶共定位(F) |
| |
In addition to MK, isopentyl diphosphate isomerase (IDI) is also the key enzyme in the terpenoids biosynthesis pathway. According to the structure of IDI, L141, Y195, and W256 are near the active pocket, forming hydrogen bonds with the neighboring β-phosphates of IPP[15]. Notably, F195 and C256, which are nonpolar amino acids, were positioned in proximity to the hydrophobic group of IPP, indirectly expanding the hydrophobic range of the active pocket[15]. The mutated IDI enzyme (Y195F and W256C) exhibited a 10% decrease in Km and the kcat was 28% improved compared to the original IDI[15]. Consequently, the enzyme activity of IDI mutation L141H-Y195F-W256C was remarkably enhanced by 2.53-fold[15]. This increase in enzyme activity corresponded to a substantial boost (2.10-fold) in lycopene production[15]. These findings highlight the significance of the identified mutations within the active pocket, leading to improved enzyme activity and ultimately improve the production of terpenoids.
1.1.2 Changing the preference of cofactorsCofactors remain vital elements for the activity of terpenoid synthases. By manipulating the cofactors involved in enzymatic reactions, researchers have successfully modulated the catalytic efficiency of terpenoid synthases, ultimately leading to increased terpene yields.
For instance, in a study focused on the production of artemisinin, a valuable antimalarial compound, cofactor changing was employed to improve the activity of amorpha-4, 11-diene synthase (ADS)[16]. Through rational enzyme modification, the NADPH-binding sites of ADS were modified, resulting in an increased preference for NADH as the cofactor[16]. This alteration led to a 3-fold enhancement in artemisinic acid production, ultimately contributing to the improved overall yield of artemisinin[16]. Similarly, in another investigation, boosting the biosynthesis of taxadiene, a precursor of the anticancer drug paclitaxel, cofactor engineering was utilized[17]. By introducing mutations in the taxadiene synthase (TS) enzyme, the researchers successfully altered its cofactor preference from NADPH to NADH. This modification resulted in a 2.4-fold increase in taxadiene production, thus facilitating the subsequent bioconversion steps towards paclitaxel production[17].
Some sesquiterpene synthases have been shown to utilize Mg2+ as a cofactor, while conifer monoterpene synthases, like pinene synthase, exhibit a preference for Mn2+[18]. However, the cytosolic concentration of Mn2+ in E. coli is relatively low (around 10 μmol/L), whereas the concentration of Mg2+ is much higher (10−20 mmol/L). This difference creates a limitation for pinene production. In an interesting investigation, scientists conducted a single round of mutagenesis and screening on pinene synthase[18]. Surprisingly, the resulting variant of the enzyme demonstrated a shift in its metal dependency from Mn2+ to Mg2+. This alteration enabled a remarkable increase in pinene productivity, with approximately 140 mg/L achieved in E. coli cultures[18]. These examples highlight the potential of cofactor preference changes in terpenoid synthases to enhance terpene production.
1.1.3 Enhancing solubilityIn synthetic metabolic pathways, achieving optimal function and efficiency of enzymes is crucial for successful implementation. When it comes to expressing non-native proteins, such as enzymes derived from plants involved in terpenoid synthesis, in microbial systems, the in vivo properties of these enzymes, such as solubility and stability, often face challenges[19]. To overcome these challenges, researchers commonly use codon optimization and promoter engineering to enhance protein expression levels. When expressing membrane bound proteins in E. coli, most often a high amount of inclusion bodies are formed. This is due to the fact that E. coli lacks inner organelles and thus the native protein cannot be properly incorporated into membranes. Consequently, the hydrophobic regions are displayed and agglomerate to inclusion bodies[9]. To further improve the in vivo properties of enzymes, truncation of the membrane anchor region or replacement with solubilizing leader peptides from highly expressed heterologous P450s has been explored. These approaches have been shown to enhance the catalytic performance of enzymes and overcome issues related to low solubility and expression levels[9].
One successful example of enhancing the expression and function of enzymes is demonstrated by the replacement of the N-terminal membrane anchor region of CYP76AH4 from Rosmarinus officinalis, to improve soluble expression of heterologous protein CYP76AH4 by replacing this region with the leader sequence MAKKTSSKGK, researchers were able to achieve functional expression of CYP76AH4 in the cytosol of E. coli[20]. This modification enabled CYP76AH4 to catalyze the oxidation of abietatriene, resulting in the production of the diterpenoid ferruginol[20]. Another example illustrates the enhancement of 8-cadinene hydroxylase (CAH) expression by modifying the N-terminal membrane anchor. Researchers replaced the predicted transmembrane sequences of the native CAH with N-terminal sequences from CAH derived from other species, leading to a significant 5-fold improvement in the activity of 8-cadinene hydroxylase[21]. Overall, by employing strategies such as modifying of N-terminal membrane anchor sequences and adding leading peptides to target enzymes, researchers can overcome challenges related to enzyme expression levels and solubility.
1.2 Enhancing enzyme specificity 1.2.1 Substrate specificityProteins can acquire novel functions and specificities in response to various environmental changes through molecular evolution by changing substrate range and catalytic activity. Erg20 facilitates the biochemical reaction that transforms IPP and DMAPP into FPP, which is a key precursor molecule in the biosynthesis of various terpenoids. Based on the structure of FPP synthase (Gallus gallus FPS1), the researchers utilized a homology modeling approach to construct a model of Erg20[22]. From this model, a specific set of residues was identified that form the active site cavity, namely Y95, F96, L97, Q168, and Y201[22]. These residues were selected due to their potential to create additional space within the cavity, which is necessary to accommodate FPP as a co-substrate for GGPP production, by introducing mutations to these residues, either individually or in combination, it would be possible to modify the active site cavity and enable it to accommodate FPP (Figure 3C)[22]. The resulting Erg20 variant with GGPP synthetic activity, co-expressed with 8-hydroxy copalyl diphosphate synthase and cultured in shake flasks, enhanced sclareol production levels by approximately 70-fold[23].
In another study, bulky residues in the middle of the catalytic site were removed and added to the prenyl diphosphate synthases through an enzyme-substrate docking technique to utilize longer and shorter substrates[24]. From results the engineering of prenyltransferases have been shown to accept unfamiliar substrates affording a series of novel terpenoids[24]. Besides this, a novel terpenes C11 containing 11-carbons was synthesized using a GPP methyltransferase, through which monoterpene precursor was methylated to give 2-methyl-GPP (2meGPP), and consumption efficacy of 2meGPP through various monoterpene synthases increased to 25%, showed the use of synthetic substrates and enhanced the chemical space of terpenes[24]. In an experimental set, ratio of C11 terpenes further increased in yeast by substituting tyrosine instead of phenylalanine at the 571st position in SfCinS1, results show about 6-fold enhancement in the affinity for 2meGPP and total catalytic efficiency remained the same[24]. These findings underscore the potential of harnessing enzymatic engineering to generate novel terpenoids.
1.2.2 Product specificityEnzymes possess the ability to selectively catalyze specific chemical reactions and produce a particular product, which is referred to as product specificity. Enzymes exhibit varying degrees of product specificity, with some enzymes are highly specific and catalyzing only a single reaction, while others may catalyze multiple related reactions. Product specificity of enzymes is determined by the precise arrangement of their active sites, which allows them to bind and interact with specific substrates and facilitate the formation of specific products[25]. From Abies grandis, an important example is γ-humulene synthase using FPP as a substrate and produces more than fifty types of sesquiterpenes[26]. Researchers used the crystal structure of 5-epi-aristolochene synthase as a model to check the plasticity of γ-humulene synthase by identifying 19 different active-site residues based on homology structure[26]. Results showed by application of saturation mutagenesis to specific residues, four residues significantly affected catalytic product specificity: W315, M447, S484, and Y566, located at the active site[26]. Mutations to these residues shifted the relative selectivity (the amount of one product relative to another product) by 100- to 1 000-fold[26]. These results suggest that plasticity residues could significantly drive molecular evolution, and most of the substitutions in plasticity residues additively affect protein functions.
By modifying product specificity, the chemical space of terpenoids can be greatly improved. It is not only reflected in the modification of terpenoid cyclase, but also has the same effect on other terpenoid enzymes. Through targeted mutations, researchers successfully modified the active site of Pinus taeda abietadiene oxidase. These mutations involved hydrophobic residues of varying sizes, which influenced the type of products produced by the enzyme. Remarkably, these modifications altered the overall pocket volume without affecting its chemical properties. As a result, novel compounds including 18-hydroxy miltiradiene, 19-hydroxy miltiradiene, and 19-hydroxy manool were discovered. Notably, these compounds had not been previously reported in any other P450 enzyme[27].
In addition, terpenoid synthase product specific modification strategy can be applied to control the ratios of components in plant essential oils (PEOs) during biotechnological production (Figure 3D). The focus of the study was on analyzing the catalytic reaction pathways of two santalene synthases, SaSSy and SanSyn, utilizing advanced multiscale simulations. During the investigation, they identified a critical residue, F441, within the SanSyn enzyme that influences the conformational dynamics of intermediates[28]. This residue specifically facilitates direct deprotonation through the involvement of the general base T298, resulting in the predominant production of α-santalene[28]. To broaden the range of synthesized products, they introduced a mutation at this residue, generating a new mutant enzyme named SanSyn F441V[28]. This mutant enzyme exhibited the ability to produce both α- and β-santalenes and showed a favorable product profile, with 57.2% α-santalene, 28.6% β-santalene, 6.7% epi-β-santalene, and 7.6% exo-α-bergamotene[28]. These component ratios successfully achieved a desired blend of primary essential oil (PEO) constituents, aligning well with the ISO 3518:2002 standard[28]. The successful manipulation of the enzyme through enzyme modification highlights a remarkable approach in constructing biotechnological platforms for PEOs that possess desirable product ratios. The above analysis revealed that the majority of the introduced mutations have a significant role in determining the catalytic outcome of the enzymes.
1.3 Enhancing enzyme stabilityEnzyme stability is a complex concept that refers to the capacity of enzymes to retain their three-dimensional structures, which are crucial for catalysis, in storage and operational processes[29]. This stability is influenced by a multitude of interactions, including hydrophobic bonds, hydrogen bonds, salt bridges, protein surface charge, disulfide bonds, and metal ions. Given the complexity of these interactions, enhancing enzyme stability presents a significant challenge, particularly due to the partial understanding of the structure-function relationship, which complicates the identification of suitable mutation targets.
However, advancements in enzyme engineering have led to the development of innovative strategies for enzyme stabilization[30]. Enzyme engineering techniques have successfully been employed to enhance the thermostability of isoprene synthase from Ipomoea batatas (IspSib), an enzyme previously identified as a limiting factor for isoprene production in engineered E. coli. To achieve this, scientists created a unique tripartite protein folding system called lac′-IspSib-′lac, this system linked the stability of IspSib to antibiotic ampicillin resistance, facilitating high-throughput screening of variants[31]. Through two rounds of random mutation and site-saturation mutation, three variants were identified: IspSib N397V-A476V, IspSib N397V-A476T, and IspSib N397V-A476C. These variants exhibited improved thermostability, which was confirmed through in vitro tests. The melting temperatures of N397V-A476V, N397V-A476T, and N397V-A476C were (45.1±0.9) ℃, (46.1±0.7) ℃, and (47.2±0.3) ℃, respectively. All of these values exceeded the melting temperature of the wild-type IspSib (41.5±0.4) ℃. Notably, the use of the IspSib N397V-A476T variant resulted in a significant increase in isoprene production during shake-flask fermentation. Isoprene production was boosted by 1.94-fold, reaching a level of 1.3 g/L[31] (Figure 3B). From another investigation, it has shown that Gly and Pro are significantly less mutable, compared to other amino acids in E. coli central metabolic enzymes. Gly and Pro mediated mutation of γ-humulene synthase increased the folding capacity of mutant variants at elevated temperatures. This increased the sesquiterpene production from 3.3-fold (at 20 ℃) to 220-fold (at 37 ℃), supporting the stability of enzymes in a heterologous system[32].
1.4 Enhancing mass transfer of multiple enzymesEnzyme co-localization is a straightforward approach of consecutive coordination of enzymes to further enhance catalytic efficiencies[33]. When heterologous enzymes become unable to collaborate with native enzymes as a result microbial metabolite production is reduced, this in turn, can result in the degradation, diffusion, or utilization of intermediates by competitive pathways, leading to a reduction in overall production and the release of toxic and reactive metabolites that pose a threat to the survival of the host cell[34]. To prevent the accumulation of toxic metabolites, co-localized enzymes are coordinated in a proximity, ensuring higher catalytic rates of sequential pathway enzymes[33]. The method is similar to natural enzymatic systems, in order to have enhanced metabolic products, their metabolic flux is increased by protein tunnel through direct substrate channeling[35]. In the process of terpene synthesis, the coordinated action of multiple enzymes is another key factor in improving the overall catalytic efficiency and product quality. Through enzyme engineering techniques, we can fuse enzymes or co-expressed in subcellular organelles to achieve more efficient substrate delivery and product accumulation. Substrate channeling is improved by bringing active sites closer through enzyme fusion, a straightforward approach (Figure 3E).
Production of α- and β-pinene can be improved by fusing and co-expressing enzymes. However, the stability and construction of the resulting proteins are dependent on the order of the domains. Comparing the fusion of GPPS to pinene synthase at the C and N-terminus, it was observed that fusion at the C-terminus had an unfavorable effect on pinene production. When pinene synthase and GPPS were fused in the correct order, pinene production in E. coli was enhanced by approximately 32 mg/L[36]. Therefore, it is essential to consider the order of enzyme fusion when attempting to improve terpene production[36]. Respectively, the production of β-phellandrene was significantly enhanced by a specific strategy involving fusion of β-phellandrene synthase with the β-subunit of phycocyanin, which is encoded by the endogenous cpcB gene, in Synechocystis transformants. The resulting improvement was remarkable, with a 3.2 mg/g of total dry cell weight that is almost 100-fold increase in β-phellandrene production[37]. In a similar research, the overexpression of chloramphenicol (cmR) and kanamycin (nptI) resistance genes in fusion with GPPS is investigated in Synechocystis[38]. Improved expression level of GPPS through fusion constructs, showing that both heterologous (such as nptI and cmR) or homologous (like cpcB) genes having high expressions are considered as leader sequence and play a crucial role in the enhanced expression of required gene, which is vital for expressing isoprenoid pathway genes. From subsequent studies, it has been revealed that in biotechnology for terpenoid production, enzyme fusion is considered one of the most promising approaches.
Moreover, besides fusion proteins colocalization can be further promoted by another alternative technique includes attachment of synthetic scaffolds to enzymes in a sequential and programmable order (Figure 3F). In synthetic scaffolds, desired enzymes are connected in a modular manner to interactive domains. It is found in studies that the intermediate pathway flux controls the production enzyme to scaffold ratio is found as an important factor[39]. Several scaffolds were produced synthetically with a diverse number of repeated metazoan signaling proteins, such as the PSD95/DlgA/Zo-1 (PDZ) domain, SH3 domain, and GTPase binding domain. It is suggested that through this mevalonate titer is improved up to 77-fold compared to synthetic scaffold, as the intermediate processing is higher due to enzyme colocalization[40]. The farnesene production was boosted up to 135% by enzyme scaffolding in fed-batch cultivations, showing enhanced overall farnesene compared with non-scaffolded pathway[41]. Activity loss and undesirable conformational changes may occur in scaffolded enzyme assemblies due to the dissimilar properties of individual enzyme components[41]. This drawback was overcome by examining interactions between short peptide tags comprising of the 18 amino acids RIAD peptide and 44 amino acids RIDD peptide, which originated from a kinase-anchoring protein, AMP-dependent protein kinase[42]. It is found that due to robust binding ability of RIDD and RIAD peptides at a 2:1 stoichiometry, without scaffold assemblies of modular enzymes. The interaction of both peptides was used and fused with geranylgeranyl diphosphate synthase (CrtE) and IDI to yield carotenoids in S. cerevisiae and E. coli. In E. coli, fusion of IDI with CrtE indicated an increase of 5.7-fold increase (276.3 mg/L) in the over-all carotenoid production[42]. Likewise, in S. cerevisiae, lycopene yield increases by up to 58% as a result of the IDI-CrtE complex formation, which leads to ultimate titer of 2.3 g/L in fed-batch fermentation[42]. The optimized strategies of genetic manipulation and novel enzyme fusions demonstrate the potential for improving the production of valuable compounds and also highlight the importance of exploring alternative techniques for enhancing metabolic pathways and generate desirable products.
2 Conclusion and prospectsThis study has highlighted the various enzyme engineering approaches that have been used to enhance the production of terpenoids. These approaches include improving catalytic activity, varying the selectivity of substrates or products, enhancing stability, and enhancing mass transfer of multiple enzymes. Examples of enzyme engineering strategies are presented in Table 1−4, with detailed information on enzymes involved in the MVA pathway, MEP pathway, and exogenous terpenoid synthesis pathway and their respective genetic modifications, resulting in changed enzyme function and product yield. Enzyme engineering strategies hold immense potential for the microbial biosynthesis of terpenoids, and significant advancements have been made in recent years through directed evolution and rational design approaches. However, it still has the problems of labor-intensive directed evolution and the low accuracy of rational design. Below, we provide an outlook on the application of advanced enzyme engineering technology in terpene synthesis.
| Enzyme | Substrate | Product | Mutations | Biological effects | References |
| Hydroxymethyl glutaryl-CoA synthase (HMGS) | Acetoacetyl- CoA | HMG-CoA | (C159A) | Production of mevalonate enhanced by 25-fold | [43] |
| Mevalonate kinase (MK) | Mevalonate | Mevalonate phosphate | (V13D-S148I-V301E) | Production of lycopene enhanced by 2.4-fold | [14] |
| Isopentenyl disphosphate isomerase (IDI) | IPP | DMAPP | (L141H-Y195F-W256C) | Catalytic activity enhanced by 2.53-fold | [15, 44] |
| 3-hydroxyl-3- methylglutaryl-CoA reductase (HMGR) | HMG-CoA | Mevalonate | (P200A-G206A-T239P- G319A-G352A-G417A- P428G-K474G-G495A) | Production of mevalonate enhanced by 3-fold | [32] |
| GGPP synthase (GGPPS) | GPP | GGPP | (P96C) | Production of sclareol enhanced by 70-fold | [23] |
| Deoxyxylulose 5-phosphate synthase (PtDXS) | G3P | DXP | (A147G-A352G) | Enzyme activity improved by 10.9-fold | [45] |
| 8-cadinene hydroxylase (P450-CYP76AH4) | Cadinene | 8-hydroxy cadinene | Substitution of lysine and serine residues at N-terminal region | Production of 8-hydroxycadinene enhanced by 5-fold | [21] |
| IPP: Isopentenyl pyrophosphate; HMG-CoA: 3-hydroxy-3-methylglutaryl coenzyme A; GPP: Geranyl disphosphate; G3P: Glyceraldehyde-3-phosphate; DMAPP: Dimethylallyl pyrophosphate; GGPP: Geranylgeranyl diphosphate; DXP: 1-deoxy-d-xylulose 5-phosphate. | |||||
| Enzyme | Substrate | Product | Mutations | Biological effects | References |
| 1, 8-cineole synthase (SfCinS1) |
2meGPP | C11 terpenes | (N388S-I451A) | Enhanced substrate specificity; Production of C11 terpene enhanced by 6-fold | [24] |
| Santalene synthase (SanSyn) |
FPP | Alpha, beta and epi-beta santalene | (F441V) | Mutant SanSyn F441V possesses the ability to produce both α- β- and Epi-β-santalenes | [6] |
| γ-humulene synthase | FPP | Bisabolene | (A336V-M447H- M562T) |
Enhanced product promiscuisity; Production improved up to 1 000-fold | [26] |
| Lycopene cyclase | GGPP | β-carotene | (Y27R) | Enhanced product selectivity; production of β-carotene was significantly enhanced, by 1 441-fold | [46] |
| Pentalenene synthase | FPP | Pentalenene, β-chamigrene, β-elemene, sativene | (T182A) | Enhanced product diversity; with pentalene the sativene, β-chamigrene, thujopsene-I3 and β-elemene also produced | [47] |
| 2meGPP: 2-methyl-geranyl disphosphate; FPP: Farnesyl diphosphate. | |||||
| Enzyme | Substrate | Product | Mutations | Biological effects | References |
| Isoperene synthase |
DMAPP | Isoprene | (N397V-A476T) | Thermostability optimized and production of isoprene enhanced by 1.94-fold | [31] |
| IDI | IPP | DMAPP | (L141H-Y195F-W256C) | Enhanced enzyme stability, the half-lives at 25–50 ℃ were 2.6-6 times longer. | [15] |
| τ-muurolol synthase | FPP | τ-muurolol | C terminus truncation | Remains active at up to 78 ℃ | [19] |
| Co-localization strategy | Substrate | Product | Improved production | References |
| Fusion of CrtZ and CrtW using three different sizes of flexible linkers of glycine, serine and proline | β-carotene | Astaxanthin | Production of astaxanthin improved by 1.4-fold | [48] |
| Co-localization and spatial organization of NES and FPPS | FPP | Nerolidol | Production of nerolidol enhanced by 50-fold | [49] |
| Fusion of ispA and α-farnesene synthase using two repeats of the GGGGS linker | FPP | α-farnesene | Production of α-farnesene enhanced by 1.5-fold | [50] |
| Fusion of Erg20, Bts1 and SmCPS, SmKSL using GGGS linker | IPP, DMAPP, GGPP | Diterpenoids | Production of miltiradiene enhanced by 4.4-fold | [51] |
Directed evolution has been discussed as an important technique when specific properties and structural information related to catalytic activities of enzymes are unknown[40]. However, the process of generating gene libraries with effective genetic diversification can be time- consuming, taking days or even weeks. In this regard, in vivo mutagenesis provides a promising alternative for achieving genetic diversity. A notable approach gaining attention is the continuous directed evolution of proteins in vivo, which includes techniques such as phage-assisted continuous evolution, directed evolution based on DNA polymerase mutations, and retroelement- based in vivo mutagenesis[52]. Efficient screening methods play a crucial role in the successful implementation of directed evolution technology. Recently in the field of synthetic biology has witnessed significant advancements, leading to the development of various biosensing components that are utilized in biosensor-based screening systems. These biosensors have the ability to convert changes in ligand concentrations into easily detectable output signals through transcriptional or RNA regulation. This enables the coupling of ligand concentration with fluorescence signals or cell growth, facilitating high-throughput screening capabilities. By employing biosensor based screening systems, researchers can effectively identify and select improved biocatalysts for further optimization through directed evolution[53]. Isoprene is a fundamental structural component found in terpenoids, which are a diverse class of natural compounds. It has made significant progress in this field by developing a high-throughput screening method using a whole-cell biosensor for the detection of isoprene synthase[54]. This approach provides a solid foundation for the future development of sensors that can selectively respond to terpenoids[54].
One approach to streamline experimental screening and reduce library size is through "semi-rational" prescreening. In recent years, various computational tools have been developed to identify target residues that are more likely to yield positive mutations, resulting in higher- quality libraries. These include sequence-based methods, structure-based methods, molecular dynamics (MD), and machine learning. Sequence- based methods, such as FOLDEF and PoPMuSiC 2.1, analyze protein stability, while SIFT, PANTHER, and PROVEAN assess the functional impact of amino acid substitutions. Structure-based methods, on the other hand, utilize tools like CAVER 3.0 to analyze protein structure channels, CASTp, LPC, and CSU software for protein interface analysis, and Q-SiteFinder, SITEHOUND-web, LigPlot+, and HotSpot Wizard for ligand binding site analysis and mutation hotspot prediction. Molecular dynamics (MD) simulation is a powerful technique that captures detailed information about the position and velocity of every atom in a system, providing insights to information which is unobtainable through experiments alone. MD simulation-assisted directed evolution methods have been successfully employed in the design of terpene synthases with different product specificities[55]. With the rapid development of data science, machine learning is becoming increasingly widespread in enzyme engineering. Its main purpose is to construct a sequence-function mapping through measured data and guide experiments to explore the relevance more effectively. However, to further enhance the precision and reliability of these predictions, the integration of quantum mechanics/molecular mechanics (QM/MM) techniques has become increasingly pertinent. QM/MM offers a unique blend of computational approaches, combining the accuracy of quantum mechanical calculations with the computational efficiency of molecular mechanical methods. This hybrid approach allows for a more detailed and accurate understanding of the intermolecular interactions at play, thereby leading to more reliable predictions of mutational outcomes. By incorporating QM/MM into our arsenal of computational tools, we can expect to achieve even higher-quality mutation libraries, with mutations that are not just positive but also optimized for specific desired outcomes[56]. It is believed that with the continuous development of computational tools, the directed evolution of terpene bioproduct-related enzymes will become faster and more efficient[52].
To avoid long screening process, rational design utilizing computational methods has been considered as another potential approach. Such as de novo design will become increasingly important in enzyme engineering, it is based on the physical and chemical principles of protein folding, and various structures can be designed from scratch under the guidance of computational molecular simulation without relying on existing protein structures as templates[57]. Efficient employment of these strategies, based on the structure and chemical principles of protein folding, leads to optimal terpenoids production. In the future, the identification and construction of novel enzymes involved in terpenoid biosynthesis will be facilitated by modern techniques in computational biology.
Recently, the development of self-driving laboratories has revolutionized enzyme engineering. The self-driving autonomous machines for protein landscape exploration (SAMPLE) is an advanced system that revolutionizes enzyme engineering by automating and expediting the process. It seamlessly integrates automated learning, decision-making, protein design, and experimentation to efficiently explore fitness landscapes and discover optimized proteins. This cutting-edge technology serves as a promising gateway for future research endeavors in the field[58].
With the continuous development of enzyme engineering technology, its application in terpene synthesis will become more extensive and in-depth. In the future, we are expected to use enzyme engineering technology to achieve the development of more efficient, selective, and stable terpene synthases. This will provide more bioactive substances and solutions for the fields of biomedicine, agriculture, and chemicals. At the same time, through the in-depth study of interdisciplinary research such as synthetic biology and metabolic engineering, we will be able to better understand and optimize the terpene synthesis pathways of microorganisms, achieving more precise and efficient production of terpene compounds.
| [1] |
WANG YH, XU HC, ZOU J, CHEN XB, ZHUANG YQ, LIU WL, CELIK E, CHEN GD, HU D, GAO H, WU RB, SUN PH, DICKSCHAT JS. Catalytic role of carbonyl oxygens and water in selinadiene synthase[J]. Nature Catalysis, 2022, 5: 128-135. DOI:10.1038/s41929-022-00735-0 |
| [2] |
THOLL D. Biosynthesis and biological functions of terpenoids in plants[J]. Advances in Biochemical Engineering/Biotechnology, 2015, 148: 63-106. |
| [3] |
WEHRS M, GLADDEN JM, LIU YZ, PLATZ L, PRAHL JP, MOON J, PAPA G, SUNDSTROM E, GEISELMAN GM, TANJORE D, KEASLING JD, PRAY TR, SIMMONS BA, MUKHOPADHYAY A. Sustainable bioproduction of the blue pigment indigoidine: expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides[J]. Green Chemistry, 2019, 21(12): 3394-3406. DOI:10.1039/C9GC00920E |
| [4] |
BUREAU JA, OLIVA ME, DONG YM, IGNEA C. Engineering yeast for the production of plant terpenoids using synthetic biology approaches[J]. Natural Product Reports, 2023, 40(12): 1822-1848. |
| [5] |
ZHU K, KONG J, ZHAO BX, RONG LX, LIU SQ, LU ZH, ZHANG CY, XIAO DG, PUSHPANATHAN K, FOO JL, WONG A, YU AQ. Metabolic engineering of microbes for monoterpenoid production[J]. Biotechnology Advances, 2021, 53: 107837. DOI:10.1016/j.biotechadv.2021.107837 |
| [6] |
ZHANG G, WANG H, ZHANG Z, VERSTREPEN KJ, WANG QH, DAI ZJ. Metabolic engineering of Yarrowia lipolytica for terpenoids production: advances and perspectives[J]. Critical Reviews in Biotechnology, 2022, 42(4): 618-633. |
| [7] |
HU ZH, LI HX, WENG YR, LI P, ZHANG CY, XIAO DG. Improve the production of d-limonene by regulating the mevalonate pathway of Saccharomyces cerevisiae during alcoholic beverage fermentation[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(12): 1083-1097. |
| [8] |
PHULARA SC, RAJPUT VS, MAZUMDAR B, RUNTHALA A. Metabolic and enzyme engineering for the microbial production of anticancer terpenoids[J]. Essentials of Cancer Genomic, Computational Approaches and Precision Medicine. Singapore: Springer, 2020, 237-259. |
| [9] |
XIAO H, ZHANG Y, WANG M. Discovery and engineering of cytochrome P450s for terpenoid biosynthesis[J]. Trends in Biotechnology, 2019, 37(6): 618-631. DOI:10.1016/j.tibtech.2018.11.008 |
| [10] |
STEINER K, SCHWAB H. Recent advances in rational approaches for enzyme engineering[J]. Computational and Structural Biotechnology Journal, 2012, 2: e201209010. DOI:10.5936/csbj.201209010 |
| [11] |
DIAZ JE, LIN CS, KUNISHIRO K, FELD BK, AVRANTINIS SK, BRONSON J, GREAVES J, SAVEN JG, WEISS GA. Computational design and selections for an engineered, thermostable terpene synthase[J]. Protein Science: a Publication of the Protein Society, 2011, 20(9): 1597-1606. DOI:10.1002/pro.691 |
| [12] |
GREENHAGEN BT, O'MAILLE PE, NOEL JP, CHAPPELL J. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(26): 9826-9831. |
| [13] |
BILAL M, IQBAL HMN. Tailoring multipurpose biocatalysts via protein engineering approaches: a review[J]. Catalysis Letters, 2019, 149(8): 2204-2217. DOI:10.1007/s10562-019-02821-8 |
| [14] |
CHEN HL, LIU CQ, LI MJ, ZHANG HB, XIAN M, LIU HZ. Directed evolution of mevalonate kinase in Escherichia coli by random mutagenesis for improved lycopene[J]. RSC Advances, 2018, 8(27): 15021-15028. DOI:10.1039/C8RA01783B |
| [15] |
CHEN HL, LI MJ, LIU CQ, ZHANG HB, XIAN M, LIU HZ. Enhancement of the catalytic activity of isopentenyl diphosphate isomerase (IDI) from Saccharomyces cerevisiae through random and site-directed mutagenesis[J]. Microbial Cell Factories, 2018, 17(1): 65. DOI:10.1186/s12934-018-0913-z |
| [16] |
DIETRICH JA, YOSHIKUNI Y, FISHER KJ, WOOLARD FX, OCKEY D, McPHEE DJ, RENNINGER NS, CHANG MCY, BAKER D, KEASLING JD. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450(BM3)[J]. ACS Chemical Biology, 2009, 4(4): 261-267. DOI:10.1021/cb900006h |
| [17] |
EDGAR S, LI FS, QIAO KJ, WENG JK, STEPHANOPOULOS G. Engineering of taxadiene synthase for improved selectivity and yield of a key taxol biosynthetic intermediate[J]. ACS Synthetic Biology, 2017, 6(2): 201-205. DOI:10.1021/acssynbio.6b00206 |
| [18] |
TASHIRO M, KIYOTA H, KAWAI-NOMA S, SAITO K, IKEUCHI M, IIJIMA Y, UMENO D. Bacterial production of pinene by a laboratory-evolved pinene-synthase[J]. ACS Synthetic Biology, 2016, 5(9): 1011-1020. DOI:10.1021/acssynbio.6b00140 |
| [19] |
STYLES MQ, NESBITT EA, MARR S, HUTCHBY M, LEAK DJ. Characterization of the first naturally thermostable terpene synthases and development of strategies to improve thermostability in this family of enzymes[J]. The FEBS Journal, 2017, 284(11): 1700-1711. DOI:10.1111/febs.14072 |
| [20] |
ZI JC, PETERS RJ. Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae[J]. Organic & Biomolecular Chemistry, 2013, 11(44): 7650. |
| [21] |
CHANG MCY, EACHUS RA, TRIEU W, RO DK, KEASLING JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s[J]. Nature Chemical Biology, 2007, 3: 274-277. DOI:10.1038/nchembio875 |
| [22] |
TARSHIS LC, YAN MJ, POULTER CD, SACCHETTINI JC. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-. ANG. resolution[J]. Biochemistry, 1994, 33(36): 10871-10877. DOI:10.1021/bi00202a004 |
| [23] |
IGNEA C, PONTINI M, MAFFEI ME, MAKRIS AM, KAMPRANIS SC. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase[J]. ACS Synthetic Biology, 2014, 3(5): 298-306. DOI:10.1021/sb400115e |
| [24] |
IGNEA C, PONTINI M, MOTAWIA MS, MAFFEI ME, MAKRIS AM, KAMPRANIS SC. Synthesis of 11-carbon terpenoids in yeast using protein and metabolic engineering[J]. Nature Chemical Biology, 2018, 14: 1090-1098. DOI:10.1038/s41589-018-0166-5 |
| [25] |
PAZOUKI L, NIINEMETS Ü. Multi-substrate terpene synthases: their occurrence and physiological significance[J]. Frontiers in Plant Science, 2016, 7: 1019. |
| [26] |
YOSHIKUNI Y, FERRIN TE, KEASLING JD. Designed divergent evolution of enzyme function[J]. Nature, 2006, 440: 1078-1082. DOI:10.1038/nature04607 |
| [27] |
IGNEA C, IOANNOU E, GEORGANTEA P, LOUPASSAKI S, TRIKKA FA, KANELLIS AK, MAKRIS AM, ROUSSIS V, KAMPRANIS SC. Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast[J]. Metabolic Engineering, 2015, 28: 91-103. DOI:10.1016/j.ymben.2014.12.001 |
| [28] |
ZHA WL, ZHANG F, SHAO JQ, MA XM, ZHU JX, SUN PH, WU RB, ZI JC. Rationally engineering santalene synthase to readjust the component ratio of sandalwood oil[J]. Nature Communications, 2022, 13: 2508. DOI:10.1038/s41467-022-30294-8 |
| [29] |
LIU Q, XUN GH, FENG Y. The state-of-the-art strategies of protein engineering for enzyme stabilization[J]. Biotechnology Advances, 2019, 37(4): 530-537. DOI:10.1016/j.biotechadv.2018.10.011 |
| [30] |
DANIEL RM, DANSON MJ, HOUGH DW, LEE CK, COWAN DA. Enzyme Stability and Activity at High Temperatures[M]. Nova Science Publishers, 2008.
|
| [31] |
LI MJ, YANG RM, GUO J, LIU M, YANG JM. Optimization of IspSib stability through directed evolution to improve isoprene production[J]. Applied and Environmental Microbiology, 2023, 89(10): e0121823. DOI:10.1128/aem.01218-23 |
| [32] |
YOSHIKUNI Y, DIETRICH JA, NOWROOZI FF, BABBITT PC, KEASLING JD. Redesigning enzymes based on adaptive evolution for optimal function in synthetic metabolic pathways[J]. Chemistry & Biology, 2008, 15(6): 607-618. |
| [33] |
DALETOS G, STEPHANOPOULOS G. Protein engineering strategies for microbial production of isoprenoids[J]. Metabolic Engineering Communications, 2020, 11: e00129. DOI:10.1016/j.mec.2020.e00129 |
| [34] |
LEE H, DeLOACHE WC, DUEBER JE. Spatial organization of enzymes for metabolic engineering[J]. Metabolic Engineering, 2012, 14(3): 242-251. DOI:10.1016/j.ymben.2011.09.003 |
| [35] |
PRÖSCHEL M, DETSCH R, BOCCACCINI AR, SONNEWALD U. Engineering of metabolic pathways by artificial enzyme channels[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 168. |
| [36] |
SARRIA S, WONG B, GARCÍA MARTÍN H, KEASLING JD, PERALTA-YAHYA P. Microbial synthesis of pinene[J]. ACS Synthetic Biology, 2014, 3(7): 466-475. DOI:10.1021/sb4001382 |
| [37] |
FORMIGHIERI C, MELIS A. A phycocyanin·phellandrene synthase fusion enhances recombinant protein expression and β-phellandrene (monoterpene) hydrocarbons production in Synechocystis (cyanobacteria)[J]. Metabolic Engineering, 2015, 32: 116-124. DOI:10.1016/j.ymben.2015.09.010 |
| [38] |
BETTERLE N, MELIS A. Heterologous leader sequences in fusion constructs enhance expression of geranyl diphosphate synthase and yield of β-phellandrene production in cyanobacteria (Synechocystis)[J]. ACS Synthetic Biology, 2018, 7(3): 912-921. DOI:10.1021/acssynbio.7b00431 |
| [39] |
TIPPMANN S, ANFELT J, DAVID F, RAND JM, SIEWERS V, UHLÉN M, NIELSEN J, HUDSON EP. Affibody scaffolds improve sesquiterpene production in Saccharomyces cerevisiae[J]. ACS Synthetic Biology, 2017, 6(1): 19-28. DOI:10.1021/acssynbio.6b00109 |
| [40] |
LI CY, ZHANG RH, WANG J, WILSON LM, YAN YJ. Protein engineering for improving and diversifying natural product biosynthesis[J]. Trends in Biotechnology, 2020, 38(7): 729-744. DOI:10.1016/j.tibtech.2019.12.008 |
| [41] |
JIA F, NARASIMHAN B, MALLAPRAGADA S. Materials-based strategies for multi-enzyme immobilization and co-localization: a review[J]. Biotechnology and Bioengineering, 2014, 111(2): 209-222. DOI:10.1002/bit.25136 |
| [42] |
KANG W, MA T, LIU M, QU JL, LIU ZJ, ZHANG HW, SHI B, FU S, MA JC, LAI LTF, HE SC, QU JN, AU SWN, KANG BH, YU LAU WC, DENG ZX, XIA J, LIU TG. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux[J]. Nature Communications, 2019, 10: 4248. DOI:10.1038/s41467-019-12247-w |
| [43] |
MARSAFARI M, XU P. Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica[J]. Metabolic Engineering Communications, 2020, 10: e00121. DOI:10.1016/j.mec.2019.e00121 |
| [44] |
LIANG ZJ, ZHI H, FANG ZX, ZHANG PZ. Genetic engineering of yeast, filamentous fungi and bacteria for terpene production and applications in food industry[J]. Food Research International, 2021, 147: 110487. DOI:10.1016/j.foodres.2021.110487 |
| [45] |
BANERJEE A, PREISER AL, SHARKEY TD. Correction: engineering of recombinant poplar deoxy-d-xylulose-5-phosphate synthase (PtDXS) by site-directed mutagenesis improves its activity[J]. PLoS One, 2016, 11(10): e0165028. DOI:10.1371/journal.pone.0165028 |
| [46] |
MA YS, LIU N, GREISEN P, LI JB, QIAO KJ, HUANG SW, STEPHANOPOULOS G. Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica[J]. Nature Communications, 2022, 13: 572. DOI:10.1038/s41467-022-28277-w |
| [47] |
LIU HS, FANG SB, ZHAO L, MEN X, ZHANG HB. A single active-site mutagenesis confers enhanced activity and/or changed product distribution to a pentalenene synthase from Streptomyces sp. PSKA01[J]. Bioengineering, 2023, 10(3): 392. |
| [48] |
NOGUEIRA M, ENFISSI EMA, WELSCH R, BEYER P, ZURBRIGGEN MD, FRASER PD. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: a new tool for engineering ketocarotenoids[J]. Metabolic Engineering, 2019, 52: 243-252. |
| [49] |
CHEAH LC, LIU L, PLAN MR, PENG BY, LU ZY, SCHENK G, VICKERS CE, SAINSBURY F. Product profiles of promiscuous enzymes can be altered by controlling in vivo spatial organization[J]. Advanced Science, 2023, 10(32): e2303415. |
| [50] |
WANG CL, YOON SH, JANG HJ, CHUNG YR, KIM JY, CHOI ES, KIM SW. Metabolic engineering of Escherichia coli for α-farnesene production[J]. Metabolic Engineering, 2011, 13(6): 648-655. |
| [51] |
ZHOU YJ, GAO W, RONG QX, JIN GJ, CHU HY, LIU WJ, YANG W, ZHU ZW, LI GH, ZHU GF, HUANG LQ, ZHAO ZK. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production[J]. Journal of the American Chemical Society, 2012, 134(6): 3234-3241. |
| [52] |
WANG YJ, XUE P, CAO MF, YU TH, LANE ST, ZHAO HM. Directed evolution: methodologies and applications[J]. Chemical Reviews, 2021, 121(20): 12384-12444. |
| [53] |
ZHANG F, ZENG T, WU RB. QM/MM modeling aided enzyme engineering in natural products biosynthesis[J]. Journal of Chemical Information and Modeling, 2023, 63(16): 5018-5034. |
| [54] |
HWANG HG, YE DY, JUNG GY. Biosensor-guided discovery and engineering of metabolic enzymes[J]. Biotechnology Advances, 2023, 69: 108251. |
| [55] |
KIM SK, KIM SH, SUBHADRA B, WOO SG, RHA E, KIM SW, KIM H, LEE DH, LEE SG. A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli[J]. ACS Synthetic Biology, 2018, 7(10): 2379-2390. |
| [56] |
LOU TT, LI AN, XU HC, PAN JF, XING BY, WU RB, DICKSCHAT JS, YANG DH, MA M. Structural insights into three sesquiterpene synthases for the biosynthesis of tricyclic sesquiterpenes and chemical space expansion by structure-based mutagenesis[J]. Journal of the American Chemical Society, 2023, 145(15): 8474-8485. |
| [57] |
BAKER D. What has de novo protein design taught us about protein folding and biophysics?[J]. Protein Science, 2019, 28(4): 678-683. |
| [58] |
RAPP JT, BREMER BJ, ROMERO PA. Self-driving laboratories to autonomously navigate the protein fitness landscape[J]. Nature Chemical Engineering, 2024, 1(1): 97-107. |
 2024, Vol. 40
2024, Vol. 40