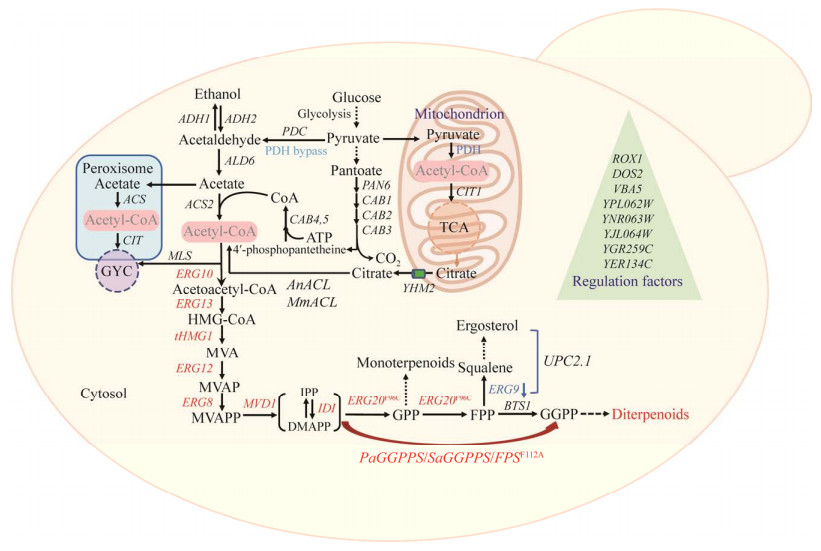
| [1] |
HU ZM, LIU XY, TIAN M, MA Y, JIN BL, GAO W, CUI GH, GUO J, HUANG LQ. Recent progress and new perspectives for diterpenoid biosynthesis in medicinal plants. Medicinal Research Reviews, 2021, 41(6): 2971-2997. DOI:10.1002/med.21816
|
|
| [2] |
ZHANG Y, JIANG PX, YE M, KIM SH, JIANG C, LÜ JX. Tanshinones: sources, pharmacokinetics and anti-cancer activities. International Journal of Molecular Sciences, 2012, 13(12): 13621-13666. DOI:10.3390/ijms131013621
|
|
| [3] |
WEAVER BA. How taxol/paclitaxel kills cancer cells. Molecular Biology of the Cell, 2014, 25(18): 2677-2681. DOI:10.1091/mbc.e14-04-0916
|
|
| [4] |
XIANG YL, YANG N, GUO ZT, ZHOU L, GUO JJ, HU M. Cost-effectiveness analysis of ginkgolide injection in the treatment of ischemic stroke based on a randomized clinical trial. The Journal of Alternative and Complementary Medicine, 2021, 27(4): 331-341. DOI:10.1089/acm.2020.0455
|
|
| [5] |
TU LC, SU P, ZHANG ZR, GAO LH, WANG JD, HU TY, ZHOU JW, ZHANG YF, ZHAO YJ, LIU Y, SONG YD, TONG YR, LU Y, YANG J, XU C, JIA MR, PETERS RJ, HUANG LQ, GAO W. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nature Communications, 2020, 11: 971. DOI:10.1038/s41467-020-14776-1
|
|
| [6] |
KINGSTON DGI, JAGTAP PG, YUAN H, SAMALA L. The chemistry of taxol and related taxoids[M]// Progress in the Chemistry of Organic Natural Products/Fortschritte der Chemie organischer Naturstoffe. Vienna: Springer Vienna, 2002: 53-225.
|
|
| [7] |
NICOLAOU KC, DAI WM, GUY RK. Chemistry and biology of taxol. Angewandte Chemie International Edition in English, 1994, 33(1): 15-44. DOI:10.1002/anie.199400151
|
|
| [8] |
ZHOU ZL, YANG YX, DING J, LI YC, MIAO ZH. Triptolide: structural modifications, structure-activity relationships, bioactivities, clinical development and mechanisms. Natural Product Reports, 2012, 29(4): 457. DOI:10.1039/c2np00088a
|
|
| [9] |
ZENG F, WANG W, GUAN SH, CHENG CR, YANG M, AVULA B, KHAN I, GUO DA. Simultaneous quantification of 18 bioactive constituents in Tripterygium wilfordii using liquid chromatography- electrospray ionization-mass spectrometry. Planta Medica, 2013, 79(9): 797-805. DOI:10.1055/s-0032-1328596
|
|
| [10] |
YU RM, ZHU JH, ZENG ZH, CHEN LL, WEN W. Biosynthesis pathways of ginkgolides. Pharmacognosy Reviews, 2013, 7(1): 47. DOI:10.4103/0973-7847.112848
|
|
| [11] |
PADDON CJ, WESTFALL PJ, PITERA DJ, BENJAMIN K, FISHER K, McPHEE D, LEAVELL MD, TAI A, MAIN A, ENG D, POLICHUK DR, TEOH KH, REED DW, TREYNOR T, LENIHAN J, JIANG H, FLECK M, BAJAD S, DANG G, DENGROVE D, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature, 2013, 496(7446): 528-532. DOI:10.1038/nature12051
|
|
| [12] | |
|
| [13] |
ZHANG J, HANSEN LG, GUDICH O, VIEHRIG K, LASSEN LMM, SCHRÜBBERS L, ADHIKARI KB, RUBASZKA P, CARRASQUER-ALVAREZ E, CHEN L, D'AMBROSIO V, LEHKA B, HAIDAR AK, NALLAPAREDDY S, GIANNAKOU K, LALOUX M, ARSOVSKA D, JØRGENSEN MAK, CHAN LJG, KRISTENSEN M, et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature, 2022, 609(7926): 341-347. DOI:10.1038/s41586-022-05157-3
|
|
| [14] |
GAO K, ZHA WL, ZHU JX, ZHENG C, ZI JC. A review: biosynthesis of plant-derived labdane-related diterpenoids. Chinese Journal of Natural Medicines, 2021, 19(9): 666-674. DOI:10.1016/S1875-5364(21)60100-0
|
|
| [15] |
SHAO J, SUN YW, LIU HL, WANG Y. Pathway elucidation and engineering of plant-derived diterpenoids. Current Opinion in Biotechnology, 2021, 69: 10-16. DOI:10.1016/j.copbio.2020.08.007
|
|
| [16] |
WANG CL, LIWEI M, PARK JB, JEONG SH, WEI GY, WANG YJ, KIM SW. Microbial platform for terpenoid production: Escherichia coli and yeast. Frontiers in Microbiology, 2018, 9: 2460. DOI:10.3389/fmicb.2018.02460
|
|
| [17] |
PARAMASIVAN K, MUTTURI S. Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae. Critical Reviews in Biotechnology, 2017, 37(8): 974-989. DOI:10.1080/07388551.2017.1299679
|
|
| [18] |
WANG ZB, ZHANG RB, YANG Q, ZHANG JT, ZHAO YX, ZHENG YN, YANG JM. Recent advances in the biosynthesis of isoprenoids in engineered Saccharomyces cerevisiae[M]//Advances in Applied Microbiology. Amsterdam: Elsevier, 2021: 1-35.
|
|
| [19] |
NIELSEN J. Yeast systems biology: model organism and cell factory. Biotechnology Journal, 2019, 14(9): 1800421. DOI:10.1002/biot.201800421
|
|
| [20] | |
|
| [21] |
BIAN GK, YUAN YJ, TAO H, SHI XF, ZHONG XF, HAN YC, FU S, FANG CX, DENG ZX, LIU TG. Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6. Biotechnology Journal, 2017, 12(4): 1600697. DOI:10.1002/biot.201600697
|
|
| [22] |
KIM J, BAIDOO EEK, AMER B, MUKHOPADHYAY A, ADAMS PD, SIMMONS BA, LEE TS. Engineering Saccharomyces cerevisiae for isoprenol production. Metabolic Engineering, 2021, 64: 154-166. DOI:10.1016/j.ymben.2021.02.002
|
|
| [23] |
LV XM, XU HM, YU HW. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Applied Microbiology and Biotechnology, 2013, 97(6): 2357-2365. DOI:10.1007/s00253-012-4485-2
|
|
| [24] |
AJIKUMAR PK, XIAO WH, TYO KEJ, WANG Y, SIMEON F, LEONARD E, MUCHA O, PHON TH, PFEIFER B, STEPHANOPOULOS G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science, 2010, 330(6000): 70-74. DOI:10.1126/science.1191652
|
|
| [25] |
KEASLING JD. Synthetic biology and the development of tools for metabolic engineering. Metabolic Engineering, 2012, 14(3): 189-195. DOI:10.1016/j.ymben.2012.01.004
|
|
| [26] |
OHTO C, MURAMATSU M, OBATA S, SAKURADANI E, SHIMIZU S. Overexpression of the gene encoding HMG-CoA reductase in Saccharomyces cerevisiae for production of prenyl alcohols. Applied Microbiology and Biotechnology, 2009, 82(5): 837-845. DOI:10.1007/s00253-008-1807-5
|
|
| [27] |
POLAKOWSKI T, STAHL U, LANG C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Applied Microbiology and Biotechnology, 1998, 49(1): 66-71. DOI:10.1007/s002530051138
|
|
| [28] |
ZHANG K, DING J. In vitro anticancer effects of levopimaric acid in cisplatin-resistant human lung carcinoma are mediated via autophagy, ROS-mediated mitochondrial dysfunction, cell apoptosis and modulation of ERK/MAPK/JNK signalling pathway. Journal of the Balkan Union of Oncology, 2020, 25(1): 248-254.
|
|
| [29] |
LIU T, ZHANG CB, LU WY. Heterologous production of levopimaric acid in Saccharomyces cerevisiae. Microbial Cell Factories, 2018, 17(1): 1-10. DOI:10.1186/s12934-017-0850-2
|
|
| [30] |
MEADOWS AL, HAWKINS KM, TSEGAYE Y, ANTIPOV E, KIM Y, RAETZ L, DAHL RH, TAI AN, MAHATDEJKUL-MEADOWS T, XU L, ZHAO LS, DASIKA MS, MURARKA A, LENIHAN J, ENG D, LENG JS, LIU CL, WENGER JW, JIANG HX, CHAO L, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature, 2016, 537(7622): 694-697. DOI:10.1038/nature19769
|
|
| [31] |
IGNEA C, TRIKKA FA, KOURTZELIS I, ARGIRIOU A, KANELLIS AK, KAMPRANIS SC, MAKRIS AM. Positive genetic interactors of HMG2 identify a new set of genetic perturbations for improving sesquiterpene production in Saccharomyces cerevisiae. Microbial Cell Factories, 2012, 11(1): 1-16. DOI:10.1186/1475-2859-11-1
|
|
| [32] |
CAO X, YU W, CHEN Y, YANG S, ZHAO ZK, NIELSEN J, LUAN HW, ZHOU YJ. Engineering yeast for high-level production of diterpenoid sclareol. Metabolic Engineering, 2023, 75: 19-28. DOI:10.1016/j.ymben.2022.11.002
|
|
| [33] |
NOWROUZI B, LI RA, WALLS LE, D'ESPAUX L, MALCı K, LIANG LG, JONGUITUD-BORREGO N, LERMA-ESCALERA AI, MORONES-RAMIREZ JR, KEASLING JD, RIOS-SOLIS L. Enhanced production of taxadiene in Saccharomyces cerevisiae. Microbial Cell Factories, 2020, 19(1): 1-12. DOI:10.1186/s12934-019-1269-8
|
|
| [34] |
ASADOLLAHI MA, MAURY J, SCHALK M, CLARK A, NIELSEN J. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 2010, 106(1): 86-96.
|
|
| [35] |
HU TY, ZHOU JW, TONG YR, SU P, LI XL, LIU Y, LIU N, WU XY, ZHANG YF, WANG JD, GAO LH, TU LC, LU Y, JIANG ZQ, ZHOU YJ, GAO W, HUANG LQ. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metabolic Engineering, 2020, 60: 87-96. DOI:10.1016/j.ymben.2020.03.011
|
|
| [36] |
ENGELS B, DAHM P, JENNEWEIN S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards taxol (paclitaxel) production. Metabolic Engineering, 2008, 10(3/4): 201-206.
|
|
| [37] |
DAI ZB, LIU Y, HUANG LQ, ZHANG XL. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnology and Bioengineering, 2012, 109(11): 2845-2853. DOI:10.1002/bit.24547
|
|
| [38] |
IGNEA C, TRIKKA FA, NIKOLAIDIS AK, GEORGANTEA P, IOANNOU E, LOUPASSAKI S, KEFALAS P, KANELLIS AK, ROUSSIS V, MAKRIS AM, KAMPRANIS SC. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metabolic Engineering, 2015, 27: 65-75. DOI:10.1016/j.ymben.2014.10.008
|
|
| [39] |
WEI PP, ZHANG CB, BIAN XK, LU WY. Metabolic engineering of Saccharomyces cerevisiae for heterologous carnosic acid production. Frontiers in Bioengineering and Biotechnology, 2022, 10: 916605. DOI:10.3389/fbioe.2022.916605
|
|
| [40] |
XU YM, WANG XL, ZHANG CY, ZHOU X, XU XH, HAN LY, LV XQ, LIU YF, LIU S, LI JH, DU GC, CHEN J, LEDESMA-AMARO R, LIU L. De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts. Nature Communications, 2022, 13: 3040. DOI:10.1038/s41467-022-30826-2
|
|
| [41] | |
|
| [42] |
GÁSPÁR ME, CSERMELY P. Rigidity and flexibility of biological networks. Briefings in Functional Genomics, 2012, 11(6): 443-456. DOI:10.1093/bfgp/els023
|
|
| [43] |
CHEN Y, SIEWERS V, NIELSEN J. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS One, 2012, 7(8): e42475. DOI:10.1371/journal.pone.0042475
|
|
| [44] |
NIELSEN J. Synthetic biology for engineering acetyl coenzyme A metabolism in yeast. mBio, 2014, 5(6): e02153.
|
|
| [45] |
SHIBA Y, PARADISE EM, KIRBY J, RO DK, KEASLING JD. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metabolic Engineering, 2007, 9(2): 160-168. DOI:10.1016/j.ymben.2006.10.005
|
|
| [46] |
WEGNER SA, CHEN JM, IP SS, ZHANG YF, DUGAR D, AVALOS JL. Engineering acetyl-CoA supply and ERG9 repression to enhance mevalonate production in Saccharomyces cerevisiae. Journal of Industrial Microbiology and Biotechnology, 2021, 48(9/10): kuab050.
|
|
| [47] |
de JONG BW, SHI SB, SIEWERS V, NIELSEN J. Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microbial Cell Factories, 2014, 13(1): 1-10. DOI:10.1186/1475-2859-13-1
|
|
| [48] |
KOZAK BU, van ROSSUM HM, LUTTIK MAH, AKEROYD M, BENJAMIN KR, WU L, de VRIES S, DARAN JM, PRONK JT, van MARIS AJA. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. mBio, 2014, 5(5): e01696-14.
|
|
| [49] |
EVANS CT, SCRAGG AH, RATLEDGE C. A comparative study of citrate efflux from mitochondria of oleaginous and non-oleaginous yeasts. European Journal of Biochemistry, 2005, 130(1): 195-204. DOI:10.1111/j.1432-1033.1983.tb07136.x
|
|
| [50] |
EVANS CT, SCRAGG AH, RATLEDGE C. Reguladtion of citrate efflux from mitochondria oleaginou and non-oleaginous yeasts by adenine nucleotides. European Journal of Biochemistry, 1983, 132(3): 609-615. DOI:10.1111/j.1432-1033.1983.tb07407.x
|
|
| [51] |
RODRIGUEZ S, DENBY CM, van VU T, BAIDOO EEK, WANG G, KEASLING JD. ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microbial Cell Factories, 2016, 15: 48. DOI:10.1186/s12934-016-0447-1
|
|
| [52] |
YU T, ZHOU YJ, HUANG MT, LIU QL, PEREIRA R, DAVID F, NIELSEN J. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell, 2018, 174(6): 1549-1558.e14. DOI:10.1016/j.cell.2018.07.013
|
|
| [53] |
OLZHAUSEN J, GRIGAT M, SEIFERT L, ULBRICHT T, SCHÜLLER HJ. Increased biosynthesis of acetyl-CoA in the yeast Saccharomyces cerevisiae by overexpression of a deregulated pantothenate kinase gene and engineering of the coenzyme A biosynthetic pathway. Applied Microbiology and Biotechnology, 2021, 105(19): 7321-7337. DOI:10.1007/s00253-021-11523-4
|
|
| [54] |
ZHANG SS, YANG W, CHEN H, LIU B, LIN BX, TAO Y. Metabolic engineering for efficient supply of acetyl-CoA from different carbon sources in Escherichia coli. Microbial Cell Factories, 2019, 18(1): 1-11. DOI:10.1186/s12934-018-1049-x
|
|
| [55] |
SUN YW, CHEN Z, WANG GY, LV HJ, MAO YP, MA K, WANG Y. De novo production of versatile oxidized kaurene diterpenes in Escherichia coli. Metabolic Engineering, 2022, 73: 201-213. DOI:10.1016/j.ymben.2022.08.001
|
|
| [56] |
ZHOU YJ, GAO W, RONG QX, JIN GJ, CHU HY, LIU WJ, YANG W, ZHU ZW, LI GH, ZHU GF, HUANG LQ, ZHAO ZK. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. Journal of the American Chemical Society, 2012, 134(6): 3234-3241. DOI:10.1021/ja2114486
|
|
| [57] |
杨薇, 周雍进, 刘武军, 沈宏伟, 赵宗保. 构建酿酒酵母工程菌合成香紫苏醇. 生物工程学报, 2013, 29(8): 1185-1192. YANG W, ZHOU YJ, LIU WJ, SHEN HW, ZHAO ZK. Engineering Saccharomyces cerevisiae for sclareol production. Chinese Journal of Biotechnology, 2013, 29(8): 1185-1192 (in Chinese). DOI:10.13345/j.cjb.2013.08.004
|
|
| [58] |
SCHALK M, PASTORE L, MIRATA MA, KHIM S, SCHOUWEY M, DEGUERRY F, PINEDA V, ROCCI L, DAVIET L. Toward a biosynthetic route to sclareol and amber odorants. Journal of the American Chemical Society, 2012, 134(46): 18900-18903. DOI:10.1021/ja307404u
|
|
| [59] |
REIDER APEL A, D'ESPAUX L, WEHRS M, SACHS D, LI RA, TONG GJ, GARBER M, NNADI O, ZHUANG W, HILLSON NJ, KEASLING JD, MUKHOPADHYAY A. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Research, 2017, 45(1): 496-508. DOI:10.1093/nar/gkw1023
|
|
| [60] |
TOKUHIRO K, MURAMATSU M, OHTO C, KAWAGUCHI T, OBATA S, MURAMOTO N, HIRAI M, TAKAHASHI H, KONDO A, SAKURADANI E, SHIMIZU S. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Applied and Environmental Microbiology, 2009, 75(17): 5536-5543. DOI:10.1128/AEM.00277-09
|
|
| [61] |
KILDEGAARD KR, ARNESEN JA, ADIEGO-PÉREZ B, RAGO D, KRISTENSEN M, KLITGAARD AK, HANSEN EH, HANSEN J, BORODINA I. Tailored biosynthesis of gibberellin plant hormones in yeast. Metabolic Engineering, 2021, 66: 1-11. DOI:10.1016/j.ymben.2021.03.010
|
|
| [62] |
LEONARD E, AJIKUMAR PK, THAYER K, XIAO WH, MO JD, TIDOR B, STEPHANOPOULOS G, PRATHER KLJ. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(31): 13654-13659. DOI:10.1073/pnas.1006138107
|
|
| [63] |
GUO J, ZHOU YJ, HILLWIG ML, SHEN Y, YANG L, WANG YJ, ZHANG XN, LIU WJ, PETERS RJ, CHEN XY, ZHAO ZK, HUANG LQ. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(29): 12108-12113. DOI:10.1073/pnas.1218061110
|
|
| [64] |
LU HZ, LI FR, SÁNCHEZ BJ, ZHU ZM, LI G, DOMENZAIN I, MARCIŠAUSKAS S, ANTON PM, LAPPA D, LIEVEN C, BEBER ME, SONNENSCHEIN N, KERKHOVEN EJ, NIELSEN J. Author correction: a consensus S. cerevisiae metabolic model yeast8 and its ecosystem for comprehensively probing cellular metabolism . Nature Communications, 2020, 11: 5443. DOI:10.1038/s41467-020-19358-9
|
|
| [65] |
YUAN SF, YI XN, JOHNSTON TG, ALPER HS. De novo resveratrol production through modular engineering of an Escherichia coli- Saccharomyces cerevisiae co-culture. Microbial Cell Factories, 2020, 19(1): 1-12. DOI:10.1186/s12934-019-1269-8
|
|
| [66] |
DZANAEVA L, KRUK B, RUCHALA J, NIELSEN J, SIBIRNY A, DMYTRUK K. The role of peroxisomes in xylose alcoholic fermentation in the engineered Saccharomyces cerevisiae. Cell Biology International, 2020, 44(8): 1606-1615. DOI:10.1002/cbin.11353
|
|
| [67] |
MARTINS D, NGUYEN D, ENGLISH AM. Ctt1 catalase activity potentiates antifungal azoles in the emerging opportunistic pathogen Saccharomyces cerevisiae. Scientific Reports, 2019, 9: 9185. DOI:10.1038/s41598-019-45070-w
|
|
| [68] |
TEMPLE MD, PERRONE GG, DAWES IW. Complex cellular responses to reactive oxygen species. Trends in Cell Biology, 2005, 15(6): 319-326. DOI:10.1016/j.tcb.2005.04.003
|
|
| [69] | |
|
| [70] |
KREN A, MAMNUN YM, BAUER BE, SCHÜLLER C, WOLFGER H, HATZIXANTHIS K, MOLLAPOUR M, GREGORI C, PIPER P, KUCHLER K. War1p, a novel transcription factor controlling weak acid stress response in yeast. Molecular and Cellular Biology, 2003, 23(5): 1775-1785. DOI:10.1128/MCB.23.5.1775-1785.2003
|
|
| [71] |
MARTÍNEZ-MONTAÑÉS F, RIENZO A, POVEDA-HUERTES D, PASCUAL-AHUIR A, PROFT M. Activator and repressor functions of the Mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae. Eukaryotic Cell, 2013, 12(5): 636-647. DOI:10.1128/EC.00037-13
|
|
| [72] |
DELAHODDE A, DELAVEAU T, JACQ C. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Molecular and Cellular Biology, 1995, 15(8): 4043-4051. DOI:10.1128/MCB.15.8.4043
|
|
| [73] |
PEREIRA R, MOHAMED ET, RADI MS, HERRGÅRD MJ, FEIST AM, NIELSEN J, CHEN Y. Elucidating aromatic acid tolerance at low pH in Saccharomyces cerevisiae using adaptive laboratory evolution. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(45): 27954-27961. DOI:10.1073/pnas.2013044117
|
|
| [74] |
RONG-MULLINS X, AYERS MC, SUMMERS M, GALLAGHER JEG. Transcriptional profiling of Saccharomyces cerevisiae reveals the impact of variation of a single transcription factor on differential gene expression in 4NQO, fermentable, and nonfermentable carbon sources. G3 Genes| Genomes| Genetics, 2018, 8(2): 607-619.
|
|
| [75] |
LI XW, WANG YY, LI G, LIU QL, PEREIRA R, CHEN Y, NIELSEN J. Metabolic network remodelling enhances yeast's fitness on xylose using aerobic glycolysis. Nature Catalysis, 2021, 4(9): 783-796. DOI:10.1038/s41929-021-00670-6
|
|
| [76] |
PAUMI CM, CHUK M, SNIDER J, STAGLJAR I, MICHAELIS S. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiology and Molecular Biology Reviews, 2009, 73(4): 577-593. DOI:10.1128/MMBR.00020-09
|
|
| [77] |
LATA PANWAR S, PASRIJA R, PRASAD R. Membrane homoeostasis and multidrug resistance in yeast. Bioscience Reports, 2008, 28(4): 217-228. DOI:10.1042/BSR20080071
|
|
| [78] |
BANERJEE A, RAHMAN H, PRASAD R, GOLIN J. How fungal multidrug transporters mediate hyper resistance through DNA amplification and mutation. Molecular Microbiology, 2022, 118(1/2): 3-15.
|
|
| [79] |
DENBY CM, IM JH, YU RC, PESCE CG, BREM RB. Negative feedback confers mutational robustness in yeast transcription factor regulation. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(10): 3874-3878. DOI:10.1073/pnas.1116360109
|
|
| [80] |
MONTAÑÉS FM, PASCUAL-AHUIR A, PROFT M. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Molecular Microbiology, 2011, 79(4): 1008-1023. DOI:10.1111/j.1365-2958.2010.07502.x
|
|
| [81] |
TRIKKA FA, NIKOLAIDIS A, ATHANASAKOGLOU A, ANDREADELLI A, IGNEA C, KOTTA K, ARGIRIOU A, KAMPRANIS SC, MAKRIS AM. Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microbial Cell Factories, 2015, 14: 60. DOI:10.1186/s12934-015-0246-0
|
|
| [82] |
ZHOU K, QIAO KJ, EDGAR S, STEPHANOPOULOS G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nature Biotechnology, 2015, 33(4): 377-383. DOI:10.1038/nbt.3095
|
|
| [83] |
CHEAH LC, STARK T, ADAMSON LSR, ABIDIN RS, LAU YH, SAINSBURY F, VICKERS CE. Artificial self-assembling nanocompartment for organizing metabolic pathways in yeast. ACS Synthetic Biology, 2021, 10(12): 3251-3263. DOI:10.1021/acssynbio.1c00045
|
|
| [84] |
丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进. 合成生物学重要研究方向进展. 合成生物学, 2020, 1(1): 7-28. DING MZ, LI BZ, WANG Y, XIE ZX, LIU D, YUAN YJ. Significant research progress in synthetic biology. Synthetic Biology Journal, 2020, 1(1): 7-28 (in Chinese).
|
|
| [85] |
KARIG DK. Cell-free synthetic biology for environmental sensing and remediation. Current Opinion in Biotechnology, 2017, 45: 69-75. DOI:10.1016/j.copbio.2017.01.010
|
|
| [86] |
HAFNER J, PAYNE J, MOHAMMADIPEYHANI H, HATZIMANIKATIS V, SMOLKE C. A computational workflow for the expansion of heterologous biosynthetic pathways to natural product derivatives. Nature Communications, 2021, 12: 1760. DOI:10.1038/s41467-021-22022-5
|
|
 2023, Vol. 39
2023, Vol. 39