中国科学院微生物研究所、中国微生物学会主办
文章信息
- 果士婷, 刘盼, 王钰
- GUO Shiting, LIU Pan, WANG Yu
- 1, 3-丙二醇的微生物合成:C6-C3-C1原料体系的转变
- Microbial production of 1, 3-propanediol: a transition of feedstocks from C6 to C3 and C1 carbon sources
- 生物工程学报, 2024, 40(8): 2371-2385
- Chinese Journal of Biotechnology, 2024, 40(8): 2371-2385
- 10.13345/j.cjb.240153
-
文章历史
- Received: February 27, 2024
- Accepted: June 4, 2024
- Published: June 6, 2024
2. 国家合成生物技术创新中心, 天津 300308
2. National Center of Technology Innovation for Synthetic Biology, Tianjin 300308, China
1, 3-丙二醇(1, 3-propanediol, 1, 3-PDO)是一种重要的大宗化学品,在食品、医药、化妆品、润滑剂等领域有着广泛的应用[1]。由于1, 3-PDO的1号和3号碳原子上各有一个羟基,因此常用于聚酯、聚氨酯等聚合物的生产,其中应用最广泛的为合成纤维新材料聚对苯二甲酸丙二醇酯(polytrimethylene terephthalate, PTT)[2],这种可降解聚酯材料因其优异的拉伸和回弹特性而被广泛使用[3],加速了市场对1, 3-PDO的需求。预计到2027年,全球1, 3-PDO市场规模将高达14.43亿美元[4]。近年来,我国出台多项政策,支持生物基特别是非粮生物基1, 3-PDO及其下游产品PTT的开发。1, 3-PDO的生产最初是基于化学合成路线,包括由壳牌公司开发的环氧乙烷氢甲酰化加氢工艺和由德固赛公司开发的丙烯醛水合加氢工艺,然而苛刻的反应条件、昂贵的催化剂及底物毒性等问题,使得这两种生产方法逐渐淡出市场[5]。与化学法相比,利用可再生资源通过微生物发酵法制备1, 3-PDO的反应条件温和,工艺环保,更加绿色可持续,也因此被广泛研究并已实现工业化。
受到粮食短缺和气候变化等因素影响,微生物发酵法合成1, 3-PDO的原料体系在不断改变和优化,首先由粮食原料葡萄糖逐渐转变为非粮原料甘油,之后进一步转变为更加可持续的非粮C1原料。作为最常应用的生物制造原料,淀粉水解生产的C6糖——葡萄糖最早被用作微生物发酵生产1, 3-PDO的原料[6-8]。2003年,杜邦公司开发了一株工程化大肠杆菌(Escherichia coli),能够以葡萄糖为底物高产1, 3-PDO,并于2006年实现工业化生产,这也是首个微生物发酵法生产1, 3-PDO的工业化应用[9]。虽然目前以葡萄糖为原料生产1, 3-PDO方法已十分成熟,但随着世界粮食需求不断增长,基于葡萄糖等粮食原料的工业化生产不仅面临成本上涨的压力,还存在“与人争粮”的风险。以生物柴油副产物甘油这种非粮的C3原料生产1, 3-PDO受到越来越多研究者的关注,并已逐渐实现商业化应用。2021年,METEX NØØVISTA公司实现了以甘油为底物的生物基1, 3-PDO商业化生产[10]。近年来,我国张家港美景荣化学工业有限公司、苏州苏震生物工程有限公司、广东清大智兴生物技术有限公司等企业也已完成甘油生物发酵生产1, 3-PDO的技术开发并进一步完成产业布局[11]。尽管甘油发酵生产1, 3-PDO的技术已逐步成熟,但受生物柴油产业的影响,甘油价格波动较大。此外,1, 3-PDO作为高还原性物质,平均碳原子还原程度为5.22,高于葡萄糖的4.00和甘油的4.67[12],因此一定比例的碳源必须通过氧化过程产生足够能量供给1, 3-PDO合成,导致了葡萄糖和甘油合成1, 3-PDO的理论转化率较低。葡萄糖、甘油等生物质的碳分子最终来源于CO2,因此,以CO2或甲醇等CO2衍生物为原料的生物合成技术受到越来越多的关注。甲醇是一种高能量密度的非粮C1原料,来源广泛、价格低廉,平均碳原子还原程度为6.00,高于葡萄糖和甘油,也高于目标产物1, 3-PDO[12],因此适用于1, 3-PDO生产。目前研究人员已在实验室规模下探索了以甲醇等C1原料为底物生产1, 3-PDO的方法。
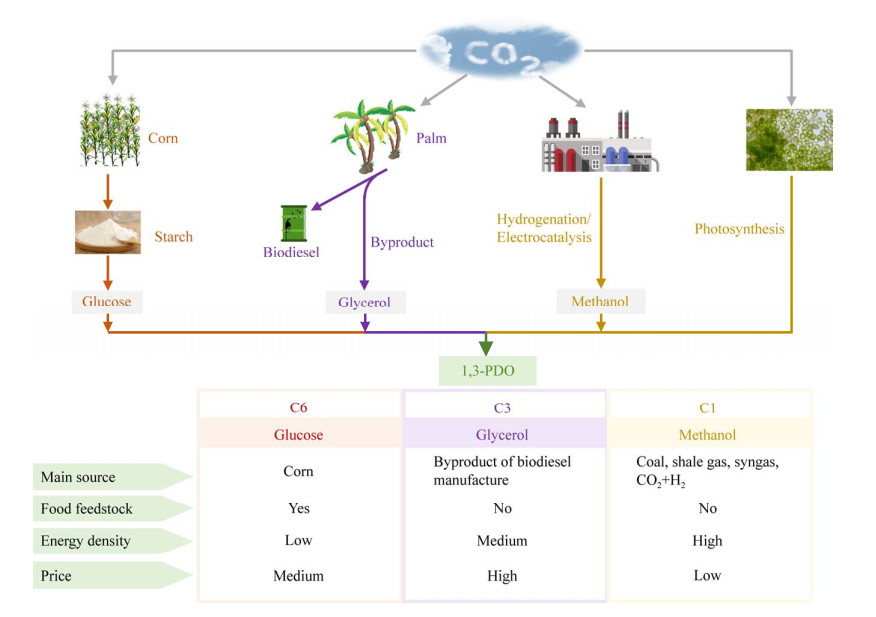
针对微生物发酵法生产1, 3-PDO,国内外多位研究者已从微生物菌种、代谢途径和发酵条件等角度进行了较为详尽的综述[2, 9-10, 13]。本文重点关注1, 3-PDO生物合成的原料体系变化趋势,综述近年来由C6、C3和C1原料合成1, 3-PDO的代表性成果,讨论不同原料体系下提升1, 3-PDO合成效率的工程化策略,并对该领域的原料体系转变进行梳理和展望(图 1)。
|
| 图 1 1, 3-PDO合成的C6、C3和C1原料体系及其优缺点 Fig. 1 C6, C3, and C1 feedstocks and their advantages and disadvantages for 1, 3-PDO synthesis. |
| |
葡萄糖是构成淀粉的主要单糖,稳定成熟的玉米深加工行业使得葡萄糖供应量大、价格相对稳定,因此葡萄糖是目前使用最为广泛的生物质原料。作为一种结构简单的单糖,葡萄糖可被自然界大多数微生物快速摄取,并以之为碳源和能源进行生长代谢和能量转化,这些特性使得葡萄糖成为许多工业发酵过程的首选底物。然而,暂时未发现可直接代谢葡萄糖合成1, 3-PDO的天然微生物,因此研究者尝试开发工程化菌株利用葡萄糖生产1, 3-PDO。
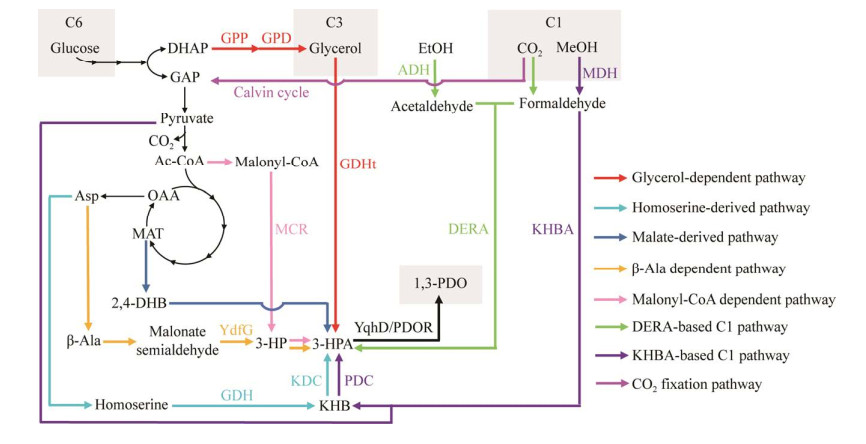
自然界中分别存在可代谢葡萄糖合成甘油和可代谢甘油合成1, 3-PDO的微生物,为打通葡萄糖合成1, 3-PDO的路线,需要在一种微生物底盘细胞中整合葡萄糖至甘油和甘油至1, 3-PDO的生物合成模块。在甘油合成模块中,葡萄糖首先经过糖酵解途径产生磷酸二羟丙酮(dihydroxylacetone phosphate, DHAP),后者再经去磷酸化和还原反应合成甘油(图 2)。在1, 3-PDO合成模块中,甘油经过脱水酶和脱氢酶的催化最终生成1, 3-PDO。该方法最具代表性的案例为2003年杜邦公司开发的工程化E. coli菌株[9],其主要的工程化策略为表达酿酒酵母(Saccharomyces cerevisiae)来源的甘油-3-磷酸脱氢酶基因gpd1和甘油-3-磷酸磷酸酶基因gpp2,以引入甘油合成模块;表达来源于天然1, 3-PDO合成菌种肺炎克雷伯氏菌(Klebsiella pneumoniae)的dha操纵子,以引入1, 3-PDO合成模块;敲除磷酸烯醇式丙酮酸(phosphoenolpyruvate, PEP)依赖的葡萄糖磷酸转移酶系统(PEP-dependent glucose phosphotransferase system, PTS),上调非PTS葡萄糖运输系统,减少PEP的消耗,下调甘油醛-3-磷酸脱氢酶,减少进入三羧酸(tricarboxylic acid, TCA)循环的碳代谢流,促进葡萄糖更多地转化为甘油。改造后的工程菌株能够以葡萄糖为底物生产高达135.0 g/L的1, 3-PDO,转化率为0.51 g/g葡萄糖,这也是目前报道的微生物发酵法生产1, 3-PDO的最高产量[9]。近期,Li等[14]开发了一株工程化的谷氨酸棒杆菌(Corynebacterium glutamicum),该菌株天然拥有丙酮酸-草酰乙酸-PEP途径,无须考虑PTS系统的PEP消耗问题。通过引入S. cerevisiae来源的甘油合成途径,并对自身特有及外源引入的甘油合成途径进行优化;整合来源于K. pneumoniae的二醇脱水酶pdu操纵子及来源于E. coli的NADPH-依赖的乙醇脱氢酶基因yqhD,引入1, 3-PDO合成模块;减少有毒中间产物3-羟基丙醛(3-hydroxypropionaldehyde, 3-HPA)和副产物3-羟基丙酸(3-hydroxypropionate, 3-HP)的积累;改造NADPH产生和消耗途径等方式实现了1, 3-PDO的高效生产,获得的重组菌株能够以葡萄糖为底物生产110.4 g/L的1, 3-PDO,转化率为0.42 g/g葡萄糖[14]。也有研究者以天然可转化甘油合成1, 3-PDO的菌株为底盘,引入甘油合成模块实现以葡萄糖为原料的1, 3-PDO生产。Lama等[7]以天然具有甘油合成1, 3-PDO途径的K. pneumoniae为底盘细胞,引入从DHAP到甘油的外源途径,构建了一株以葡萄糖合成1, 3-PDO的工程菌株,并对其进行碳代谢和能量代谢的改造,主要包括敲除PTS系统以解除碳分解代谢物阻遏,敲除副产物生成途径及甘油分泌途径,突变电子传递链中关键蛋白二氢硫酰脱氢酶和TCA循环中柠檬酸合成酶,并缺失编码NADH脱氢酶的ndh基因,从而调节TCA循环和电子传递链等,得到的工程化菌株以葡萄糖为底物可生产61.9 g/L 1, 3-PDO,转化率为0.54 g/g葡萄糖。
|
| 图 2 C6、C3和C1原料合成1, 3-PDO的主要代谢途径 Fig. 2 The main metabolic pathways for synthesizing 1, 3-PDO from C6, C3, and C1 feedstocks. DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde-3-phosphate; Ac-CoA, acetyl-coenzyme A; Asp, Aspartate; β-Ala, β-alanine; OAA, oxaloacetate; MAT, malate; 2, 4-DHB, 2, 4-dihydroxybutyrate; 3-HPA, 3-hydroxypropionaldehyde; 3-HP, 3-hydroxypropionate; KHB, 2-keto-4-hydroxybutyrate; MeOH, methanol; EtOH, ethanol; 1, 3-PDO, 1, 3-propanediol; GPD, glycerol 3-phosphate dehydrogenase; GPP, glycerol-3-phosphate phosphatase; MCR, malonyl-CoA reductase; KDC, branched-chain alpha-keto acid decarboxylase; GDHt, glycerol dehydratase; YdfG, 3-hydroxy acid dehydrogenase; ADH, alcohol dehydrogenase; MDH, NAD-dependent methanol dehydrogenase; DERA, deoxyribose-5-phosphate aldolase; KHBA, 2-keto-4- hydroxybutyrate aldolase; YqhD, NADPH-dependent alcohol dehydrogenase; PDOR, 1, 3-propanediol oxidoreductase; GDH, glutamate dehydrogenase; PDC, pyruvate decarboxylase. |
| |
此外,研究者还设计了不以甘油为中间代谢物的1, 3-PDO人工合成途径(图 2)。其中一种途径为高丝氨酸衍生途径,在该途径中高丝氨酸被氨基酸转移酶或氨基酸脱氢酶催化转化为2-酮基-4-羟基丁酸(2-keto-4-hydroxybutyrate, KHB),然后在α-酮酸脱羧酶的催化下进一步转化为3-HPA,最后3-HPA被还原为1, 3-PDO[15-17]。Zhang等[17]在E. coli中共表达突变的丝氨酸氨基转移酶SerCR42W/R77W、丙酮酸脱羧酶和NADPH-依赖的乙醇脱氢酶,获得的重组E. coli可通过补料分批发酵生产3.0 g/L 1, 3-PDO。另一种非天然途径为苹果酸衍生途径,该途径分为2, 4-二羟基丁酸(2, 4-dihydroxybutyrate, 2, 4-DHB)合成模块和1, 3-PDO合成模块,在2, 4-DHB合成模块中,苹果酸通过苹果酸激酶、苹果酸半醛脱氢酶和醛脱氢酶的催化生成2, 4-DHB,随后,1, 3-PDO合成模块中的脱氢酶、α-酮酸脱羧酶和醇脱氢酶将2, 4-DHB转化为1, 3-PDO[6, 18]。Frazão等[6]利用苹果酸衍生途径构建了一株工程E. coli,但是以葡萄糖为底物仅可生产7.6 mg/L 1, 3-PDO,产量有待提升。3-HP衍生途径是另一种1, 3-PDO的人工合成途径,该途径以3-HP为前体物质,通过3-羟基丙酰CoA合酶、醛脱氢酶和醇脱氢酶的催化,最终生成1, 3-PDO,基于该途径,研究者分别设计了整合丙二酰-CoA生成途径和β-丙氨酸生成途径的合成方法,以葡萄糖为底物分别可产生8.0 g/L和11.2 g/L 1, 3-PDO[19-20]。上述非天然1, 3-PDO合成途径为1, 3-PDO的生产提供了更加经济的新思路。然而,目前使用这些人工途径的菌种的产量较低,需要寻求更加高效、特异的蛋白元件进一步提高生产水平。
目前,研究者开发了多种以葡萄糖为原料合成1, 3-PDO的方法(表 1),其中杜邦公司[9]开发的工程化E. coli和Li等[14]开发的工程化C. glutamicum已实现工业化应用。然而,葡萄糖的平均碳原子还原度低于1, 3-PDO,导致以葡萄糖为底物生产1, 3-PDO时,需要产生更高氧化态的副产物或完全氧化产生CO2才能维持整体的电子平衡,这也导致以葡萄糖为底物合成1, 3-PDO的理论得率下降[21]。此外,我国粮食需求不断增长,加上资源环境对粮食生产的约束,用于大规模生物制造的糖基底物供应有限,利用非粮原料生产1, 3-PDO受到研究者的广泛关注。
| Feedstock | Strain | Main engineering strategy | Titer | Yield | Productivity | References |
| Glucose | Escherichia coli | Expression of gpd1, gpp2, dha operon, yqhD, galP, and glk; ΔglpK, ΔgldA, ΔptsHI, Δcrr, Δedd-eda, ΔmgsA, Δpta-ackA, Δndh, ΔarcA; downregulation of gapA | 135.0 g/L | 0.51 g/g glucose | 3.5 g/(L·h) | [9] |
| Glucose | Corynebacterium glutamicum | Expression of pduCEDGH, yqhD, gpd1, gpp2; ΔldhA: : hdpA-gldA, ΔpoxB: : hdpA-gldA, Δamn: : pntAB, Δpyk, ΔaldA, ΔadhA, Δppc, Δzwf; downregulation of gapA | 110.4 g/L | 0.42 g/g glucose | 2.3 g/(L·h) | [14] |
| Glucose | Klebsiella pneumoniae | Expression of gdp1, gpp2, glf, dhaB, gdrAB, dhaT; ΔptsG, ΔglpF, ΔldhA, ΔadhE, ΔfrdA, ΔpflB, Δedd-eda, ΔmgsA, Δpta-ackA, ΔpoxB, Δmdh, Δndh, ΔbudB; gltAR164K, lpdAE324G | 61.9 g/L | 0.54 g/g glucose | 0.9 g/(L·h) | [7] |
| Glucose | Escherichia coli | Expression of serCR42W/R77W, pdc, yqhD, metL, lysC; ΔthrB | 3.0 g/L | 19.30 mg/g glucose | 48.9 mg/(L·h) | [17] |
| Glucose | Escherichia coli | Expression of ppcK620S, lldV108C, kdcAV461I, yqhD | 7.6 mg/L | 0.38 mg/g glucose | 0.3 mg/(L·h) | [6] |
| Glucose | Escherichia coli | Expression of prpE, pduP and yqhD | 8.0 g/L | 0.10 g/g glucose | 0.2 g/(L·h) | [19] |
| Glucose | Escherichia coli | Expression of bpanD, banA, ydfG, gltA, pduP, yqhD, ppc; ΔlysC | 11.2 g/L | – | 0.1 g/(L·h) | [20] |
| –: Not available. | ||||||
甘油是一种常见的C3原料,具有高度官能化的分子形式,可经过氧化、还原、卤化、醚化和酯化等反应转化为一系列化学品。随着人们对可持续发展的关注,对生物柴油的需求日益增长,甘油作为生物柴油生产的主要副产物随之大量产生,若能将甘油转化为1, 3-PDO等高附加值化学品,可提升整个生物柴油产业的产品综合利用率和整体经济性。以甘油为底物生产1, 3-PDO逐渐成为研究热点,甘油也成为了目前1, 3-PDO生物合成的主要原料之一。
微生物利用甘油生产1, 3-PDO最早可追溯至1881年,Freund[22]发现巴氏梭状芽孢杆菌(Clostridium pasteuianu)可利用甘油产生1, 3-PDO。自然界的多种微生物具有代谢甘油产生1, 3-PDO的能力,如K. pneumoniae、C. pasteuianu、丁酸梭状芽孢杆菌(Clostridium butyricum)、短乳杆菌(Lactobacillus brevis)和弗氏柠檬酸杆菌(Citrobacter freundii)等[23-27]。甘油主要在厌氧或微耗氧条件下通过歧化途径转化为1, 3-PDO。通过歧化途径的氧化分支,甘油通过甘油激酶-甘油-3-磷酸脱氢酶途径或甘油脱氢酶-二羟基丙酮激酶途径被氧化为DHAP,然后进入中心碳代谢,为细胞生长提供NADH和ATP等能量,并一步产生乙酸、乙醇、乳酸、2, 3-丁二醇等副产物。通过还原分支,甘油首先在VB12依赖的甘油脱水酶的催化下生成3-HPA,然后通过1, 3-PDO氧化还原酶的催化生成1, 3-PDO,同时消耗氧化分支途径产生的NADH (图 2)。
甘油氧化分支途径为还原分支途径提供还原力,对维持胞内的氧化还原平衡至关重要,因此天然菌株以甘油为底物生产1, 3-PDO必然会伴随大量副产物的产生[28]。研究者试图通过敲除副产物合成途径或增强1, 3-PDO合成通量来减少副产物积累,提高1, 3-PDO转化率[29-32]。然而,副产物的形成参与调节细胞中的氧化还原平衡及中心碳代谢通量,一种副产物形成的减少往往会伴随其他副产物的增加,且会对菌体的生长和代谢产生影响[33-34]。因此,辅因子再生与平衡是以甘油为底物生产1, 3-PDO的重要策略。Wang等[35]采用辅因子工程策略,通过向K. pneumoniae中引入运动发酵单胞菌(Zymomonas mobilis)的Entner-Doudoroff途径并过表达可溶性吡啶核苷酸转氢酶,构建了NADH再生系统,重组菌株胞内NADH/NAD+较对照菌株提高了92.8%,该菌株可联产78.7 g/L 1, 3-PDO和32.1 g/L 2, 3-丁二醇,二醇得率为0.78 mol/mol甘油。Zhou等[36]在反硝化假单胞菌(Pseudomonas denitrificans)中失活NADH脱氢酶,调节细胞内氧化还原平衡,失活乙酸合成途径并阻断3-HP生成,通过最大化甘油向还原途径的碳流量,最小化甘油向氧化途径的流量,提高1, 3-PDO转化率。为提升甘油合成高值产物的利用率,Xin等[37]设计将甘油氧化分支的碳流重新定向到其他高附加值化学品,在综合考虑产品价值、碳守恒、氧化还原平衡、生物相容性和下游加工等因素后,构建了一株联产1, 3-PDO和光学纯乳酸的产酸克雷伯氏菌(Klebsiella oxytoca),将甘油歧化途径中氧化分支部分的通量用于合成乳酸,同时产生NADH用于支持还原分支途径中的1, 3-PDO合成。通过补料分批发酵,该菌株可联产76.2 g/L 1, 3-PDO和111.9 g/L D-乳酸,总转化率为0.95 mol/mol甘油,具有较高的原子经济性[37]。Wang等[38]在K. pneumoniae中敲除乳酸脱氢酶、乙醇脱氢酶、延胡索酸还原酶的编码基因ldhA、adhE和frdA,之后引入聚羟基丁酸(polyhydroxybutyrate, PHB)的合成途径,将乙酰-CoA引入PHB合成途径中,最终以甘油为底物生产了91.2 g/L 1, 3-PDO和2.6 g/L PHB。高温发酵是一种新兴的高效生物制造技术,高温生产过程中底物、产物的扩散速度加快,利于发酵效率的提高,同时还可以降低冷却成本和污染风险。近期,Chen等[39]首次尝试以嗜热菌为底盘菌株进行1, 3-PDO的生产,从42种嗜热菌中筛选出葡糖苷副产芽孢杆菌(Parageobacillus thermoglucosidasius)作为底盘菌株,并对其进行改造以消除副产物并提高甘油到1, 3-PDO的通量,得到的工程菌株最终可生产5.9 g/L的1, 3-PDO,产率为0.31 g/g甘油,后续可通过优化辅因子供应进一步优化生产(表 2)。
| Feedstock | Strain | Main engineering strategy | Titer | Yield | Productivity | References |
| Glycerol | Klebsiella pneumoniae | Expression of eda, edd, dhaT, pgl, zwf, udh; ΔldhA, ΔptsG | 78.7 g/L 1, 3-PDO and 32.1 g/L 2, 3-butanediol | Total conversion yield: 0.78 mol/mol glycerol | 2.5 g/(L·h) | [35] |
| Glycerol | Pseudomonas denitrificans | Expression of dhaB, gdrAB, dhaT; Δ3hpdh, Δ3hibdhIV, Δ3hibdhI, ΔaldH13, ΔnuoA, Δpta-ackA | 33.5 g/L | 0.74 g/g glycerol | 0.7 g/(L·h) | [36] |
| Glycerol | Klebsiella oxytoca | ΔbudA, ΔbudB, ΔadhE, ΔackA-pta, Δpox, ΔfrdA | 76.2 g/L 1, 3-PDO and 111.9 g/L D-lactate | Total conversion yield: 0.95 mol/mol glycerol | 2.5 g/(L·h) | [37] |
| Glycerol | Klebsiella pneumoniae | Expression of acs; ΔadhE, ΔldhA, ΔfrdA, ΔaceA | 91.2 g/L 1, 3-PDO and 2.6 g/L PHB | 0.49 g/g glycerol | 3.1 g/(L·h) | [38] |
| Glycerol | Parageobacillus thermoglucosidasius | Expression of pduCEDGH; ΔadhE, ΔldhA, ΔpflB | 5.9 g/L | 0.31 g/g glycerol | 0.1 g/(L·h) | [39] |
| Crude glycerol | Clostridium butyricum | Wild-type | 76.2 g/L | – | 2.3 g/(L·h) | [40] |
| Crude glycerol | Klebsiella pneumoniae | ΔldhA | 81.1 g/L | – | 3.4 g/(L·h) | [33] |
| –: Not available. | ||||||
上述研究中,生产1, 3-PDO的原料均为精甘油,然而精甘油的提纯过程复杂导致成本增加,直接利用生物柴油产业的副产品粗甘油为原料更具成本优势。目前也有研究者以粗甘油为原料进行1, 3-PDO生产。Wilkens等[40]筛选到一株可在粗甘油中快速生长的C. butyricum,该菌株以粗甘油为原料可生产76.2 g/L 1, 3-PDO,生产强度为2.3 g/(L·h)。Oh等[33]利用K. pneumoniae的乳酸脱氢酶缺失菌株,以粗甘油为原料通过补料分批发酵可生产81.1 g/L 1, 3-PDO,生产强度为3.4 g/(L·h) (表 2)。
甘油比葡萄糖拥有更高的平均碳原子还原程度,用于生产1, 3-PDO等高还原性化合物时有更高的理论得率。目前,我国生物制造企业的微生物发酵生产1, 3-PDO技术主要以甘油为原料。然而,作为生物柴油的主要副产物,甘油价格受生物柴油产业影响波动较大,目前相比于葡萄糖并无价格优势。开发新的、低成本的、适宜生产1, 3-PDO的非粮原料成为未来需要重点关注的研究方向。
3 C1原料合成1, 3-PDO随着生物制造产业的不断发展扩大,亟须寻找稳定、可持续和廉价的原料。同时,为了应对不断加剧的气候变化,我国正努力实现“碳达峰、碳中和”目标。在此背景下,发展基于甲醇、甲烷、CO2等非粮C1原料的生物合成技术有利于推进我国“双碳”目标达成,因而受到了广泛关注[41-45]。甲醇作为一种液体原料,易于运输和储存,来源丰富、价格低廉[46]。甲醇是煤炭和页岩气化工的初级平台产品,近年来,生物质经合成气重整生产甲醇和CO2加氢生产甲醇的工艺逐步成熟,我国甲醇产能也持续增长,已超8 300万t/年,占国际总量59%[47]。与葡萄糖、甘油等传统生物质原料相比,甲醇能量密度更高,是生产1, 3-PDO等高还原度化学品的理想原料[48-49]。
研究者已通过构建人工甲基营养菌或改造天然甲基营养菌获得了以甲醇为底物生产多种化学品的工程菌株[50-53]。目前,以甲醇为原料生产1, 3-PDO的研究主要集中在基于E. coli的人工甲基营养菌。Wang等[54]以E. coli为底盘细胞,设计了一条基于甲醇氧化产物甲醛和丙酮酸缩合反应的1, 3-PDO合成途径。首先,甲醇经过甲醇脱氢酶被氧化为甲醛,再经过KHB醛缩酶的催化与丙酮酸缩合为KHB,后者经支链α-酮酸脱羧酶和1, 3-PDO氧化还原酶催化经脱羧和还原反应生成1, 3-PDO (图 2)[54]。该途径巧妙地将甲醇整合进入中心代谢,1, 3-PDO合成途径较短,但受限于关键酶的活性和特异性,工程菌株仅生产了32.7 mg/L的1, 3-PDO,且需要葡萄糖作为共底物提供丙酮酸[54]。Meng等[55]设计了一条基于甲醇氧化产物甲醛和乙醇氧化产物乙醛缩合反应的途径。首先,甲醇和乙醇分别在甲醇脱氢酶和乙醇脱氢酶的催化下氧化为甲醛和乙醛,两者经脱氧核糖-5-磷酸醛缩酶催化缩合为3-HPA,后者再经还原反应生成1, 3-PDO (图 2)[55]。与上述研究相比,该途径不涉及脱羧反应,不产生CO2,理论碳转化率为100%,具有较大的应用潜力。但受限于甲醇脱氢酶和脱氧核糖-5-磷酸醛缩酶的低活性,以甲醇和乙醇为共底物时,1, 3-PDO产量仅为79.8 mg/L[55]。后续可通过蛋白质工程等手段创制更高效的催化元件,同时通过建立NADH平衡系统驱动甲醇和乙醇的氧化,提高1, 3-PDO的合成水平。
CO2也可以通过光合作用直接用于生物合成(图 2)。研究者以光合细菌细长聚球藻(Synechococcus elongatus)为底盘细胞,引入S. cerevisiae的甘油合成模块、K. pneumoniae的甘油脱水酶和E. coli的NADPH-依赖的乙醇脱氢酶,通过优化基因的表达水平和培养条件,工程菌种可利用CO2和光能在20 d内积累1.2 g/L 1, 3-PDO[56]。除了使用工程改造的单一菌种外,通过共培养将CO2和光能转化为甘油的自养微生物S. elongatus和将甘油转化为1, 3-PDO的异养微生物K. pneumoniae,也可以实现转化CO2生产1, 3-PDO,证明了直接利用CO2和光能在可持续生产1, 3-PDO等化学品中的潜力[57]。
尽管上述研究已实现生物转化甲醇等C1原料合成1, 3-PDO (表 3),但产量和转化率距离产业化应用仍有距离,且构建的人工甲基营养菌通常需要提供除甲醇外的辅助碳源,没有充分发挥甲醇作为生物合成原料的优势,因此仍需开发更高效的C1原料合成1, 3-PDO的工程菌株。相比于基于E. coli等平台菌株的人工甲基营养菌,巴斯德毕赤酵母(Pichia pastoris)、甲醇芽孢杆菌(Bacillus methanolicus)、扭脱甲基杆菌(Methylorubrum extorquens)等天然甲基营养菌具有更高效的C1原料利用能力,且已被用来高效生产3-羟基丙酸、氨基酸、聚羟基脂肪酸、植物天然产物等多种化学品[58-63]。利用合成生物学方法,开发可合成1, 3-PDO的天然甲基营养菌,有望实现C1原料高效合成1, 3-PDO。
| Feedstock | Strain | Main engineering strategy | Titer | Productivity | References |
| Methanol and glucose | Escherichia coli | Expression of khb, kdc, dhaT; ΔfrmA | 32.7 mg/L | 1.3 mg/(L·h) | [54] |
| Methanol and ethanol | Escherichia coli | Expression of derA, mdh2, DhaT | 79.8 mg/L | 1.9 mg/(L·h) | [55] |
| CO2 | Synechococcus elongatus | Expression of dhaB1, dhaB2, dhaB3, gdrA, gdrB, yqhD, lacI, gpd1, hor2 | 1.2 g/L | 2.5 mg/(L·h) | [56] |
| CO2 | Synechococcus elongatus Klebsiella pneumoniae |
Synechococcus elongatus expressing gpp Klebsiella pneumoniae wild-type |
40.0 mg/L | 0.3 mg/(L·h) | [57] |
工业经济的发展过度依赖化石资源,导致资源短缺、全球变暖等资源和环境方面的问题逐渐凸显。以可再生资源为原料,以生物系统为主要工具的生物制造有望建立绿色可持续的工业生产新模式。原料的可持续性是生物制造关注的重点,选择来源广泛、价格低廉,且“不与人争粮”的可再生原料是生物制造产业可持续发展的关键。以重要的生物基化学品和材料单体1, 3-PDO为例,近年来国内外市场对1, 3-PDO等生物基材料单体的需求不断扩大,而我国人均耕地资源和水资源有限,保障1, 3-PDO等生物基材料单体的可持续供应需要开辟除葡萄糖等粮食原料外的新原料体系。为此,国内重点开发了以生物柴油副产物甘油为原料的1, 3-PDO生产方法,1, 3-PDO的生产原料由最初的C6葡萄糖逐渐向C3甘油转变[11]。相较于葡萄糖,甘油可大量从生物柴油制造行业的废料中获得,将其转化为1, 3-PDO有利于提高生物柴油制造业的经济性[4]。此外,甘油较葡萄糖具有更高的平均碳原子还原程度,合成1, 3-PDO的理论转化率更高。但是受到生物柴油产业波动的影响,甘油价格不稳定,为工业化的1, 3-PDO生产带来了成本不稳定因素。为了积极应对气候变化和环境问题,我国提出“碳达峰、碳中和”目标,以甲醇、CO2等C1分子为原料合成食物、燃料、化学品等生产生活必需品是当前的研究热点。目前生物转化甲醇等C1原料合成1, 3-PDO的研究仍处于实验室研究阶段,现有的人工甲基营养菌的甲醇利用效率低,目标产物的生物合成水平低,亟待开发更高效的工程菌种。
一方面,亟须开发具有更高甲醇同化效率的人工甲基营养菌。通过在平台菌种E. coli和S. cerevisiae中整合甲醇同化途径,结合理性的代谢工程改造和适应性进化,研究者构建了利用甲醇为唯一碳源的人工甲基营养菌。但是,相比于以葡萄糖为原料的生长速度,这些人工甲基营养菌利用甲醇为碳源的生长速度仍较慢,甲醇利用效率有待提高[64-66]。另一方面,可开发利用具有高效天然甲醇利用能力的天然甲基营养菌,作为新型的生物合成底盘细胞,用于生物转化甲醇等C1原料合成高附加值化学品[67-69]。对于甲基营养酵母、B. methanolicus和M. extorquens等天然甲基营养菌的遗传和代谢机制研究已相对深入[70]。然而,B. methanolicus、M. extorquens等菌株的遗传操作工具仍然有限,基因编辑相对困难。受限于缺乏高效的基因编辑等使能技术,天然甲基营养菌的育种研究仍然主要通过经典的诱变和筛选来实现[71-72]。因此,开发适用于天然甲基营养菌的高效基因编辑和表达调控工具包,如CRISPR/Cas技术,有助于早日实现基于甲醇和CO2等C1原料的1, 3-PDO高效生产。除了开发高效的甲醇生物转化微生物底盘,还可以利用人工智能算法,计算设计具有高原子经济性、热力学和动力学可行的C1原料合成1, 3-PDO的新途径,设计创制新的生化反应和新的催化酶元件。在上述微生物底盘中组装人工合成途径,采用“设计-构建-测试-学习”的合成生物学理念,逐步创建可高效利用甲醇等C1原料合成1, 3-PDO的工程菌种。
| [1] |
GUNGORMUSLER-YILMAZ M, CICEK N, LEVIN DB, AZBAR N. Cell immobilization for microbial production of 1, 3-propanediol[J]. Critical Reviews in Biotechnology, 2016, 36(3): 482-494.
|
| [2] |
SAXENA RK, ANAND P, SARAN S, ISAR J. Microbial production of 1, 3-propanediol: recent developments and emerging opportunities[J]. Biotechnology Advances, 2009, 27(6): 895-913.
|
| [3] |
KURIAN JV. A new polymer platform for the future—Sorona® from corn derived 1, 3-propanediol[J]. Journal of Polymers and the Environment, 2005, 13(2): 159-167.
|
| [4] |
Da SILVA RUY AD, de BRITO ALVES RM, REIS HEWER TL, de AGUIAR PONTES D, GOMES TEIXEIRA LS, MAGALHÃES PONTES LA. Catalysts for glycerol hydrogenolysis to 1, 3-propanediol: a review of chemical routes and market[J]. Catalysis Today, 2021, 381: 243-253.
|
| [5] |
LIU HJ, XU YZ, ZHENG ZM, LIU DH. 1, 3-propanediol and its copolymers: research, development and industrialization[J]. Biotechnology Journal, 2010, 5(11): 1137-1148.
|
| [6] |
FRAZÃO CJR, TRICHEZ D, SERRANO-BATAILLE H, DAGKESAMANSKAIA A, TOPHAM CM, WALTHER T, FRANÇOIS JM. Construction of a synthetic pathway for the production of 1, 3-propanediol from glucose[J]. Scientific Reports, 2019, 9: 11576.
|
| [7] |
LAMA SM, SEOL E, PARK S. Development of Klebsiella pneumoniae J2B as microbial cell factory for the production of 1, 3-propanediol from glucose[J]. Metabolic Engineering, 2020, 62: 116-125.
|
| [8] |
SINGH K, AINALA SK, PARK S. Metabolic engineering of Lactobacillus reuteri DSM 20, 016 for improved 1, 3-propanediol production from glycerol[J]. Bioresource Technology, 2021, 338: 125590.
|
| [9] |
ZHU FH, LIU DH, CHEN Z. Recent advances in biological production of 1, 3-propanediol: new routes and engineering strategies[J]. Green Chemistry, 2022, 24(4): 1390-1403.
|
| [10] |
AGRAWAL D, BUDAKOTI M, KUMAR V. Strategies and tools for the biotechnological valorization of glycerol to 1, 3-propanediol: challenges, recent advancements and future outlook[J]. Biotechnology Advances, 2023, 66: 108177.
|
| [11] |
曾艳, 赵心刚, 周桔. 合成生物学工业应用的现状和展望[J]. 中国科学院院刊, 2018, 33(11): 1211-1217. ZENG Y, ZHAO XG, ZHOU J. Current situations and perspectives of industrial applications of synthetic biology[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1211-1217 (in Chinese). |
| [12] |
VILLADSEN J, NIELSEN J, LIDÉN G. Bioreaction Engineering Principles[M]. Berlin: Springer Science & Business Media, 2003.
|
| [13] |
ZHU YT, WANG YX, GAO H, WANG HX, WAN ZJ, JIANG YJ, XIN FX, ZHANG WM, JIANG M. Current advances in microbial production of 1, 3-propanediol[J]. Biofuels, Bioproducts and Biorefining, 2021, 15(5): 1566-1583.
|
| [14] |
LI ZH, DONG YF, LIU Y, CEN XC, LIU DH, CHEN Z. Systems metabolic engineering of Corynebacterium glutamicum for high-level production of 1, 3-propanediol from glucose and xylose[J]. Metabolic Engineering, 2022, 70: 79-88.
|
| [15] |
CHEN Z, GENG F, ZENG AP. Protein design and engineering of a de novo pathway for microbial production of 1, 3-propanediol from glucose[J]. Biotechnology Journal, 2015, 10(2): 284-289.
|
| [16] |
ZHONG WQ, ZHANG Y, WU WJ, LIU DH, CHEN Z. Metabolic engineering of a homoserine-derived non-natural pathway for the de novo production of 1, 3-propanediol from glucose[J]. ACS Synthetic Biology, 2019, 8(3): 587-595.
|
| [17] |
ZHANG YJ, MA CW, DISCHERT W, SOUCAILLE P, ZENG AP. Engineering of phosphoserine aminotransferase increases the conversion of L-homoserine to 4-hydroxy-2-ketobutyrate in a glycerol-independent pathway of 1, 3-propanediol production from glucose[J]. Biotechnology Journal, 2019, 14(9): e1900003.
|
| [18] |
WALTHER T, TOPHAM CM, IRAGUE R, AURIOL C, BAYLAC A, CORDIER H, DRESSAIRE C, LOZANO-HUGUET L, TARRAT N, MARTINEAU N, STODEL M, MALBERT Y, MAESTRACCI M, HUET R, ANDRÉ I, REMAUD-SIMÉON M, FRANÇOIS JM. Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2, 4-dihydroxybutyric acid[J]. Nature Communications, 2017, 8: 15828.
|
| [19] |
LI ZH, WU ZY, CEN XC, LIU Y, ZHANG Y, LIU DH, CHEN Z. Efficient production of 1, 3-propanediol from diverse carbohydrates via a non-natural pathway using 3-hydroxypropionic acid as an intermediate[J]. ACS Synthetic Biology, 2021, 10(3): 478-486.
|
| [20] |
LI MD, ZHANG Y, LI JC, TAN TW. Biosynthesis of 1, 3-propanediol via a new pathway from glucose in Escherichia coli[J]. ACS Synthetic Biology, 2023, 12(7): 2083-2093.
|
| [21] |
王宝光, 刘铭, 杜晨宇, 沈金玉, 曹竹安. 微生物法生产1, 3-丙二醇过程的代谢工程研究进展[J]. 过程工程学报, 2006, 6(1): 144-149. WANG BG, LIU M, DU CY, SHEN JY, CAO ZA. Recent developments in microbial metabolic engineering for the production of 1, 3-propanediol[J]. The Chinese Journal of Process Engineering, 2006, 6(1): 144-149 (in Chinese). DOI:10.3321/j.issn:1009-606X.2006.01.033 |
| [22] |
FREUND A. Über die bildung und darstellung von trimethylenalkohol aus glycerin[J]. Monatshefte für Chemie, 1881, 2: 636-641.
|
| [23] |
DONG XY, XIU ZL, LI S, HOU YM, ZHANG DJ, REN CS. Dielectric barrier discharge plasma as a novel approach for improving 1, 3-propanediol production in Klebsiella pneumoniae[J]. Biotechnology Letters, 2010, 32(9): 1245-1250.
|
| [24] |
CUI YL, ZHOU JJ, GAO LR, ZHU CQ, JIANG X, FU SL, GONG H. Utilization of excess NADH in 2, 3-butanediol-deficient Klebsiella pneumoniae for 1, 3-propanediol production[J]. Journal of Applied Microbiology, 2014, 117(3): 690-698.
|
| [25] |
PYNE ME, SOKOLENKO S, LIU XJ, SRIRANGAN K, BRUDER MR, AUCOIN MG, MOO-YOUNG M, CHUNG DA, CHOU CP. Disruption of the reductive 1, 3-propanediol pathway triggers production of 1, 2-propanediol for sustained glycerol fermentation by Clostridium pasteurianum[J]. Applied and Environmental Microbiology, 2016, 82(17): 5375-5388.
|
| [26] |
PRZYSTAŁOWSKA H, ZEYLAND J, SZYMANOWSKA-POWAŁOWSKA D, SZALATA M, SŁOMSKI R, LIPIŃSKI D. 1, 3-propanediol production by new recombinant Escherichia coli containing genes from pathogenic bacteria[J]. Microbiological Research, 2015, 171: 1-7.
|
| [27] |
DROŻDŻYŃSKA A, PAWLICKA J, KUBIAK P, KOŚMIDER A, PRANKE D, OLEJNIK-SCHMIDT A, CZACZYK K. Conversion of glycerol to 1, 3-propanediol by Citrobacter freundii and Hafnia alvei-newly isolated strains from the Enterobacteriaceae[J]. New Biotechnology, 2014, 31(5): 402-410.
|
| [28] |
JACOBS NJ, VANDEMARK PJ. Comparison of the mechanism of glycerol oxidation in aerobically and anaerobically grown Streptococcus faecalis[J]. Journal of Bacteriology, 1960, 79(4): 532-538.
|
| [29] |
CHEN Z, LIU HJ, LIU DH. Regulation of 3-hydroxypropionaldehyde accumulation in Klebsiella pneumoniae by overexpression of dhaT and dhaD genes[J]. Enzyme and Microbial Technology, 2009, 45(4): 305-309.
|
| [30] |
KIM DK, RATHNASINGH C, SONG H, LEE HJ, SEUNG D, CHANG YK. Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2, 3-butanediol production[J]. Journal of Bioscience and Bioengineering, 2013, 116(2): 186-192.
|
| [31] |
LIN J, ZHANG YQ, XU DF, XIANG G, JIA ZX, FU SL, GONG H. Deletion of poxB, pta, and ackA improves 1, 3-propanediol production by Klebsiella pneumoniae[J]. Applied Microbiology and Biotechnology, 2016, 100(6): 2775-2784.
|
| [32] |
SEO MY, SEO JW, HEO SY, BAEK JO, RAIRAKHWADA D, OH BR, SEO PS, CHOI MH, KIM CH. Elimination of by-product formation during production of 1, 3-propanediol in Klebsiella pneumoniae by inactivation of glycerol oxidative pathway[J]. Applied Microbiology and Biotechnology, 2009, 84(3): 527-534.
|
| [33] |
OH BR, LEE SM, HEO SY, SEO JW, KIM CH. Efficient production of 1, 3-propanediol from crude glycerol by repeated fed-batch fermentation strategy of a lactate and 2, 3-butanediol deficient mutant of Klebsiella pneumoniae[J]. Microbial Cell Factories, 2018, 17(1): 92.
|
| [34] |
ZHOU S, HUANG YH, MAO XL, LI LL, GUO CY, GAO YL, QIN QW. Impact of acetolactate synthase inactivation on 1, 3-propanediol fermentation by Klebsiella pneumoniae[J]. PLoS One, 2019, 14(4): e0200978.
|
| [35] |
WANG M, ZHOU YO, TAN TW. Cofactor engineering for enhanced production of diols by Klebsiella pneumoniae from co-substrate[J]. Biotechnology Journal, 2017, 12(11): 1700176.
|
| [36] |
ZHOU SF, LAMA SM, SANKARANARAYANAN M, PARK S. Metabolic engineering of Pseudomonas denitrificans for the 1, 3-propanediol production from glycerol[J]. Bioresource Technology, 2019, 292: 121933.
|
| [37] |
XIN B, TAO F, WANG Y, LIU HY, MA CQ, XU P. Coordination of metabolic pathways: enhanced carbon conservation in 1, 3-propanediol production by coupling with optically pure lactate biosynthesis[J]. Metabolic Engineering, 2017, 41: 102-114.
|
| [38] |
WANG WJ, YU X, WEI YJ, LEDESMA-AMARO R, JI XJ. Reprogramming the metabolism of Klebsiella pneumoniae for efficient 1, 3-propanediol production[J]. Chemical Engineering Science, 2021, 236: 116539.
|
| [39] |
CHEN ZF, LIU JQ, HAN X, MA LN, XU P, TAO F. Developing a thermophilic cell factory for high-temperature production of 1, 3-propanediol via host-mining and metabolic engineering[J]. Chem Catalysis, 2023, 3(8): 100704.
|
| [40] |
WILKENS E, RINGEL AK, HORTIG D, WILLKE T, VORLOP KD. High-level production of 1, 3-propanediol from crude glycerol by Clostridium butyricum AKR102a[J]. Applied Microbiology and Biotechnology, 2012, 93(3): 1057-1063.
|
| [41] |
HAYNES CA, GONZALEZ R. Rethinking biological activation of methane and conversion to liquid fuels[J]. Nature Chemical Biology, 2014, 10: 331-339.
|
| [42] |
BAR-EVEN A, NOOR E, FLAMHOLZ A, MILO R. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes[J]. Biochimica et Biophysica Acta, 2013, 1827(8/9): 1039-1047.
|
| [43] |
OLAH GA. Towards oil independence through renewable methanol chemistry[J]. Angewandte Chemie (International Edition), 2013, 52(1): 104-107.
|
| [44] |
DÜRRE P, EIKMANNS BJ. C1-carbon sources for chemical and fuel production by microbial gas fermentation[J]. Current Opinion in Biotechnology, 2015, 35: 63-72.
|
| [45] |
YISHAI O, GOLDBACH L, TENENBOIM H, LINDNER SN, BAR-EVEN A. Engineered assimilation of exogenous and endogenous formate in Escherichia coli[J]. ACS Synthetic Biology, 2017, 6(9): 1722-1731.
|
| [46] |
WANG Y, FAN LW, TUYISHIME P, ZHENG P, SUN JB. Synthetic methylotrophy: a practical solution for methanol-based biomanufacturing[J]. Trends in Biotechnology, 2020, 38(6): 650-666.
|
| [47] |
陈婉. 技术成熟, 已涉足企业有先行优势[J]. 环境经济, 2019(20): 20-23. CHEN W. Mature technology, enterprises that have set foot in have the first-mover advantage[J]. Environmental Economy, 2019(20): 20-23 (in Chinese). |
| [48] |
WHITAKER WB, SANDOVAL NR, BENNETT RK, FAST AG, PAPOUTSAKIS ET. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization[J]. Current Opinion in Biotechnology, 2015, 33: 165-175.
|
| [49] |
ZHANG WM, SONG M, YANG Q, DAI ZX, ZHANG SJ, XIN FX, DONG WL, MA JF, JIANG M. Current advance in bioconversion of methanol to chemicals[J]. Biotechnology for Biofuels, 2018, 11: 260.
|
| [50] |
BENNETT RK, GONZALEZ JE, WHITAKER WB, ANTONIEWICZ MR, PAPOUTSAKIS ET. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph[J]. Metabolic Engineering, 2018, 45: 75-85.
|
| [51] |
LEßMEIER L, PFEIFENSCHNEIDER J, CARNICER M, HEUX S, PORTAIS JC, WENDISCH VF. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate[J]. Applied Microbiology and Biotechnology, 2015, 99(23): 10163-10176.
|
| [52] |
BRAUTASET T, JAKOBSEN OM, JOSEFSEN KD, FLICKINGER MC, ELLINGSEN TE. Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50 ℃[J]. Applied Microbiology and Biotechnology, 2007, 74(1): 22-34.
|
| [53] |
BRAUTASET T, JAKOBSEN ØM, DEGNES KF, NETZER R, NÆRDAL I, KROG A, DILLINGHAM R, FLICKINGER MC, ELLINGSEN TE. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for L-lysine production from methanol at 50 ℃[J]. Applied Microbiology and Biotechnology, 2010, 87(3): 951-964.
|
| [54] |
WANG C, REN J, ZHOU LB, LI ZD, CHEN L, ZENG AP. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-keto-4-hydroxybutyrate and 1, 3-propanediol in Escherichia coli[J]. ACS Synthetic Biology, 2019, 8(11): 2483-2493.
|
| [55] |
MENG H, WANG C, YUAN QP, REN J, ZENG AP. An aldolase-based new pathway for bioconversion of formaldehyde and ethanol into 1, 3-propanediol in Escherichia coli[J]. ACS Synthetic Biology, 2021, 10(4): 799-809.
|
| [56] |
HIROKAWA Y, MAKI Y, HANAI T. Improvement of 1, 3-propanediol production using an engineered cyanobacterium, Synechococcus elongatus by optimization of the gene expression level of a synthetic metabolic pathway and production conditions[J]. Metabolic Engineering, 2017, 39: 192-199.
|
| [57] |
WANG Y, TAO F, NI J, LI C, XU P. Production of C3 platform chemicals from CO2 by genetically engineered cyanobacteria[J]. Green Chemistry, 2015, 17(5): 3100-3110.
|
| [58] |
LIANG WF, CUI LY, CUI JY, YU KW, YANG S, WANG TM, GUAN CG, ZHANG C, XING XH. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168.
|
| [59] |
ORITA I, NISHIKAWA K, NAKAMURA S, FUKUI T. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions[J]. Applied Microbiology and Biotechnology, 2014, 98(8): 3715-3725.
|
| [60] |
BRAUTASET T, WILLIAMS MD, DILLINGHAM RD, KAUFMANN C, BENNAARS A, CRABBE E, FLICKINGER MC. Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in L-lysine-secreting mutants[J]. Applied and Environmental Microbiology, 2003, 69(7): 3986-3995.
|
| [61] |
GAO JQ, LI YX, YU W, ZHOU YJ. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol[J]. Nature Metabolism, 2022, 4: 932-943.
|
| [62] |
GUO F, DAI ZX, PENG WF, ZHANG SJ, ZHOU J, MA JF, DONG WL, XIN FX, ZHANG WM, JIANG M. Metabolic engineering of Pichia pastoris for malic acid production from methanol[J]. Biotechnology and Bioengineering, 2021, 118(1): 357-371.
|
| [63] |
WU XY, CAI P, GAO LH, LI YX, YAO L, ZHOU YJ. Efficient bioproduction of 3-hydroxypropionic acid from methanol by a synthetic yeast cell factory[J]. ACS Sustainable Chemistry & Engineering, 2023, 11(16): 6445-6453.
|
| [64] |
ZHU TC, ZHAO TX, BANKEFA OE, LI Y. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: challenges and opportunities[J]. Biotechnology Advances, 2020, 39: 107467.
|
| [65] |
DAI ZX, GU HL, ZHANG SJ, XIN FX, ZHANG WM, DONG WL, MA JF, JIA HH, JIANG M. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae[J]. Bioresource Technology, 2017, 245(Pt B): 1407-1412.
|
| [66] |
MÜLLER JEN, MEYER F, LITSANOV B, KIEFER P, POTTHOFF E, HEUX S, QUAX WJ, WENDISCH VF, BRAUTASET T, PORTAIS JC, VORHOLT JA. Engineering Escherichia coli for methanol conversion[J]. Metabolic Engineering, 2015, 28: 190-201.
|
| [67] |
IRLA M, HEGGESET TMB, NÆRDAL I, PAUL L, HAUGEN T, LE SB, BRAUTASET T, WENDISCH VF. Genome-based genetic tool development for Bacillus methanolicus: Theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production[J]. Frontiers in Microbiology, 2016, 7: 1481.
|
| [68] |
CUI LY, WANG SS, GUAN CG, LIANG WF, XUE ZL, ZHANG C, XING XH. Breeding of methanol-tolerant Methylobacterium extorquens AM1 by atmospheric and room temperature plasma mutagenesis combined with adaptive laboratory evolution[J]. Biotechnology Journal, 2018, 13(6): e1700679.
|
| [69] |
YANG YM, CHEN WJ, YANG J, ZHOU YM, HU B, ZHANG M, ZHU LP, WANG GY, YANG S. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179.
|
| [70] |
高教琪, 周雍进. 甲醇生物转化的机遇与挑战[J]. 合成生物学, 2020, 1(2): 158-173. GAO JQ, ZHOU YJ. Advances in methanol bio-transformation[J]. Synthetic Biology Journal, 2020, 1(2): 158-173 (in Chinese). |
| [71] |
MOTOYAMA H, ANAZAWA H, KATSUMATA R, ARAKI K, TESHIBA S. Amino acid production from methanol by Methylobacillus glycogenes mutants: isolation of L-glutamic acid hyper-producing mutants from M. glycogenes strains, and derivation of L-threonine and L-lysine-producing mutants from them[J]. Bioscience, Biotechnology, and Biochemistry, 1993, 57(1): 82-87.
|
| [72] |
刘康, 乔杨怡, 张尚杰, 郭峰, 马江锋, 信丰学, 章文明, 姜岷. 甲醇生物转化合成化学品的研究进展[J]. 生物工程学报, 2023, 39(6): 2430-2448. LIU K, QIAO YY, ZHANG SJ, GUO F, MA JF, XIN FX, ZHANG WM, JIANG M. Advances in biotransformation of methanol into chemicals[J]. Chinese Journal of Biotechnology, 2023, 39(6): 2430-2448 (in Chinese). |
 2024, Vol. 40
2024, Vol. 40