Recent advances in metabolic engineering of microorganisms for production of tyrosol and its derivatives
Tyrosol is an important phenolic compound, also known as p-hydroxyphenyl ethanol, primarily found in olive oil, red wine, tea, and other foods. It is a natural product of Ligustrum, Rhodiola and olive[1-2]. It is widely used in food and medicine because of its biological activities containing anti-oxidation, anti-inflammation, and anti-fatigue. It also has pharmacological activities, including anti-cancer and neuroprotection[3-9]. In addition, the microbial synthesis of tyrosol (Table 1) has also attracted the attention of researchers of the synthetic biology industry, as tyrosol can be utilized as an intermediate to synthesize a variety of high-value compounds, such as hydroxytyrosol (Table 2) and salidroside (Table 3).
表 1 代谢工程改造大肠杆菌和酿酒酵母合成酪醇的研究现状
Table 1 Recent advances in metabolic engineering of Escherichia coli and Saccharomyces cerevisiae for the production of tyrosol
| Strains |
Pathway construction gene (↑) |
Synthetic pathway gene (↑) |
Competitive pathway gene |
Feedback inhibition node |
Global control |
Medium optimization |
Culture style |
Substrate |
Titer |
References |
| Saccharomyces cerevisiae BY4741 |
Pcaasopt |
Ecadhopt EctyrAfbr |
△pdc1 |
EctyrAfbr |
– |
– |
SF |
Glucose |
126.74 mg/g |
[10] |
| Escherichia coli BL21(DE3) |
Pcaas/Scadh6 |
– |
△crr/△ptsG/△fheA/△pykF/△feaB |
△tyrR |
– |
✔ |
SF |
Glucose/Glycerol |
2.12 g/L |
[11] |
| Escherichia coli BL21(DE3) |
Pcaas |
– |
△fheA/△feaB |
△tyrR |
– |
✔ |
SF |
Glucose |
0.53 g/L |
[12] |
| Escherichia coli BW25113 |
Mltyoopt/Pstdcopt |
– |
△feaB |
– |
– |
– |
SF |
Glucose |
0.07 g/L |
[13] |
| Escherichia coli MG1655 |
Scaro10 |
ppsA/tktA/aroE/aroD/aroBopt |
△pykF/△pykA/△feaB |
△tyrR/aroGfbr/tyrAfbr |
– |
– |
SF |
Glucose |
0.93 g/L |
[14] |
| Escherichia coli BL21(DE3) |
Scaro10 |
yahK/aroA/aroE |
△fheA/△feaB |
tyrAfbr |
– |
– |
SF |
Glucose |
0.80 g/L |
[15] |
| Escherichia coli BL21(DE3) |
Scaro10 |
– |
△feaB/△pheA |
– |
– |
✔ |
SF |
Glucose |
0.57 g/L |
[16] |
| Escherichia coli MG1655 |
Scaro10opt (five copies) |
– |
△feaB/△pykF/△pabAB/△trpE/△pheA/△tyrB |
△tyrR |
– |
✔ |
FB (5 L) |
Glucose |
3.90 g/L |
[17] |
| Escherichia coli DOPA 30N |
Scaro10F138L/D218G |
yahK |
△feaB |
– |
Quorum-sensing |
– |
FB (2 L) |
Glucose |
4.22 g/L |
[18] |
| Escherichia coli MG1655 |
Scaro10D331C |
– |
△tyrB/△fheA/△feaB |
△tyrR |
– |
– |
SF |
Glucose |
2.04 g/L |
[19] |
| Escherichia coli MG1655 |
Scaro10opt |
– |
△tyrB/△fheA/△feaB |
△tyrR |
– |
✔ |
SF |
Glucose |
1.32 g/L |
[20] |
| Escherichia coli BL21(DE3) |
Scaro10 |
– |
△fheA/△feaB |
– |
– |
✔ |
SF |
Glucose |
0.57 g/L |
[21] |
| Escherichia coli BL21(DE3) |
Scaro10/Scaro8 |
– |
– |
– |
– |
✔ |
SF |
Tyrosine |
1.20 g/L |
[21] |
| Escherichia coli BL21(DE3) |
Scfdc1/Ppsmo/Ppsoi/Slpar/Fjtal |
– |
△fheA |
aroGfbr/tyrAfbr |
– |
– |
SF |
Glucose/Glycerol |
0.09 g/L |
[22] |
| Escherichia coli BL21(DE3) |
Scfdc1/Ppsmo/Ppsoi/Slpar/Fjtal |
– |
△fheA |
aroGfbr/tyrAfbr |
– |
– |
SF |
Glucose/Glycerol |
0.55 g/L |
[22] |
| Escherichia coli MG1655(DE3) |
Scfdc1/Ppsmo/Ppsoi/Slpar/Fjtal |
– |
– |
– |
– |
– |
SF |
p-CA |
2.07 g/L |
[22] |
| Saccharomyces cerevisiae BY4741 |
Bbxfpkopt/Pcaasopt |
PahpaBCopt |
– |
aro4fbr/aro7fbr |
Glucose-response |
✔ |
SF |
Sucrose/Glycerol |
0.46 g/L |
[23] |
| Saccharomyces cerevisiae HLF‐Dα |
Bbxfpkopt/Pcaasopt |
EctyrAfbr |
△pdc1/△pha2/△trp3 |
EctyrAfbr |
– |
– |
FB (3 L) |
Glucose |
8.48 g/L |
[24] |
| Saccharomyces cerevisiae CEN.PK2-1C |
– |
rki1/tkl1/aro2/aro10 |
△pdc1/△pha2 |
aro4fbr/aro7fbr/aro3fbr |
– |
– |
SF |
Glucose |
0.70 g/L |
[25] |
| Escherichia coli BL21(DE3) |
Pmkdc/Pmadh/Pmgdh/CmaadV438G/K147V/R151E |
– |
△feaB |
– |
– |
✔ |
SF |
Tyrosine |
1.32 g/L |
[26] |
| aroE: Shikimate dehydrogenase; aroB: 3-dehydroquinate synthase; aroD: 3-dehydroquinate dehydratase; aro2: Bifunctional chorismate synthase; aro7: Chorismate mutase; aroG: DAHP synthase; aro4: Tyrosine synthesis are DAHP synthases; Bbxfpk: Phosphoketolase from Bifidobacterium breve; Cmaad: L-amino acid deaminase from Cosenzaea myxofaciens; crr: Phosphoenolpyruvate carbohydrate phosphotransferase system; EctyrA: Prephenate dehydrogenase from E. coli; Fjtal: Tyrosine ammonia-lyase from Flavobacterium johnsoniae; feaB: Phenylacetaldehyde dehydrogenase; Mltyo: Tyramine oxidase from Micrococcus luteus; Pstdc: Tyrosine decarboxylase from Papaver somniferum; Pmkdc: α-keto acid decarboxylase from P. mirabilis; Pmadh: Alcohol dehydrogenase from P. mirabilis; Pmgdh: Glucose dehydrogenase from P. mirabilis; ppsA: Phosphoenolpyruvate synthase; Pcaas: Aromatic aldehyde synthase from Petroselinum crispum; Ppsmo: Styrene monooxygenase from Pseudomonas putida; Ppsoi: Styrene oxide isomerase from Pseudomonas putida; pabB: p-aminobenzoic acid synthase component Ⅰ; pabA: p-aminobenzoic acid synthase component Ⅱ; pykF: Pyruvate kinase isozymes; pheA: Chorismate synthase; pdc1: Pyruvate decarboxylase 1; pha2: Prephenate dehydratase; rki1: Ribose-5-phosphate ketol-isomerase; Scaro10: Pyruvate decarboxylase from Saccharomyces cerevisiae; Scadh6: Alcohol dehydrogenases from Saccharomyces cerevisiae; Scfdc1: Ferulic acid decarboxylase from Saccharomyces cerevisiae; Slpar: Phenylacetaldehyde reductase from Solanum lycopersicum; Scaro8: Aromatic amino acid aminotransferase from Saccharomyces cerevisiae; tktA: Transketolase; tkl1: Transketolase; trpE: Component Ⅰ gene of anthranilate synthase; tyrB: Aromatic amino acid transaminase; tyrR: A transcriptional regulator; trp3: Prephenate dehydratase; yahK: Alcohol dehydrogenase. FB: Fed-batch fermentation; SF: Shake-flask fermentation. The "–" indicates that this metabolic engineering operation is not performed. The "✔" indicates that related metabolic engineering is performed. The "↑" indicates that related gene is overexpressed. In all the tables and figures of this review, the upper case represents the protein, and the corresponding lower case represents the gene that encodes the protein. |
表 2 代谢工程改造大肠杆菌和酿酒酵母合成羟基酪醇的研究现状
Table 2 Recent advances in metabolic engineering of Escherichia coli and Saccharomyces cerevisiae for the production of hydroxytyrosol
| Strains |
Pathway construction gene (↑) |
Synthetic pathway gene (↑) |
Competitive pathway gene |
Feedback inhibition node |
Global control |
Medium optimization |
Culture style |
Substrate |
Titer (g/L) |
References |
| Escherichia coli BL21(DE3) |
Pcaas |
hpaBC |
△fheA/△feaB |
△tyrR |
– |
✔ |
SF |
Glucose |
0.21 |
[12] |
| Saccharomyces cerevisiae BY4741 |
Bbxfpkopt/Pcaasopt |
PahpaBCopt |
– |
aro4fbr/aro7fbr |
Glucose-response |
✔ |
SF |
Sucrose/Glycerol |
0.31 |
[23] |
| Saccharomyces cerevisiae-Escherichia coli co-culture |
Bbxfpkopt/Pcaasopt |
hpaBC |
– |
aro4fbr/aro7fbr |
Glucose-response |
✔ |
SF |
Sucrose |
0.44 |
[27] |
| Escherichia coli BL21(DE3) |
Scaro10/Scadh6/Pmlaad |
hpaBC/tyrC |
△crr/△ptsG/△pheA |
△tyrR/aroGfbr |
– |
– |
SF |
Glucose/Glycerol |
3.12 |
[28] |
| Escherichia coli BL21(DE3) |
Scaro10/Scadh6/Pmlaad |
hpaBC/tyrC |
△crr/△ptsG/△pheA |
△tyrR/aroGfbr |
– |
✔ |
FB (5 L) |
Glucose/Glycerol |
9.87 |
[28] |
| Escherichia coli BW25113 |
Scaro10/Scadh6 |
hpaBC |
△feaB |
– |
– |
✔ |
SF |
Glucose/Glycerol |
0.65 |
[29] |
| Escherichia coli BW25113 |
Scaro10/Scadh6 |
hpaBC |
△feaB |
– |
– |
✔ |
SF |
Tyrosine |
1.24 |
[29] |
| Escherichia coli MG1655 |
Scaro10opt (five copies) |
hpaBC |
△poxB/△feaB/△pykF/△pabAB/△typE/△pheA/△tyrB |
△tyrR |
– |
– |
SF |
Glucose |
1.81 |
[30] |
| Escherichia coli MG1655 |
Scaro10opt (five copies) |
hpaBC |
△poxB/△feaB/△pykF/△pabA/△pabB/△typE/△pheA/△tyrB |
△tyrR |
– |
– |
FB (5 L) |
Glucose |
2.95 |
[30] |
| Escherichia coli BL21(DE3) |
Pstdcopt/Mltyoopt |
hpaBC |
△fheA/△feaB/ |
tyrAfbr/aroGfbr/△tyrR |
– |
– |
SF |
Glucose |
0.27 |
[31] |
| Escherichia coli-Escherichia coli co-culture |
Abpdcopt/Rparopt |
tyrB/hpaBC/gdh |
– |
– |
– |
✔ |
SF |
Tyrosine |
1.42 |
[32] |
| Escherichia coli MG1655 |
Scaro10opt (five copies) |
hpaBS210T/A211L/Q212E/Y282H/hpaC |
△feaB/△pykF/△pabA/△pabB/△typE/△pheA/△tyrB |
△tyrR |
– |
✔ |
SF |
Tyrosol |
7.43 |
[33] |
| Saccharomyces cerevisiae CEN.PK2-1C |
EctyrAM53I A354V/Pahpab/Echpac |
rki1/tkl1/aro2/aro10 |
△pdc1/△pha2/trp2↓ |
aro4fbr/aro7fbr/aro3fbr |
– |
– |
SF |
Glucose |
1.12 |
[34] |
| Saccharomyces cerevisiae CEN.PK2-1C |
EctyrAM53I A354V/PahpaB/EchpaC |
rki1/tkl1/aro2/aro10 |
△pdc1/△pha2/trp2↓ |
aro4fbr/aro7fbr/aro3fbr |
– |
– |
FB (5 L) |
Glucose |
6.97 |
[34] |
| Saccharomyces cerevisiae BY4743 |
EchpaBc |
– |
– |
– |
– |
– |
SF |
Tyrosine |
< 0.01 |
[35] |
| Saccharomyces cerevisiae BY4743 |
EchpaBC |
– |
– |
– |
– |
– |
SF |
Tyrosol |
< 0.01 |
[35] |
| Saccharomyces cerevisiae BY4743 |
EchpaBC |
– |
– |
aro4fbr |
– |
✔ |
SF |
Glucose |
0.38 |
[36] |
| HpaBC: 4-hydroxyphenylacetate 3-monooxygenase. The annotations for other gene abbreviations are consistent with Table 1. |
表 3 代谢工程改造大肠杆菌和酿酒酵母合成红景天苷的研究现状
Table 3 Recent advances in metabolic engineering of Escherichia coli and Saccharomyces cerevisiae for the production of salidroside
| Strains |
Pathway construction gene (↑) |
Synthetic pathway gene (↑) |
Competitive pathway gene |
Feedback inhibition node |
Global control |
Medium optimization |
Culture style |
Substrate |
Titer (g/L) |
References |
| Escherichia coli BL21(DE3) |
Pcaas Atugt85a1 |
– |
△fheA/△feaB |
△tyrR |
– |
✔ |
SF |
Glucose |
0.29 |
[12] |
| Escherichia coli MG1655 |
Scaro10 Rsugt73b6opt |
ppsA/tktA/aroE/aroD/aroBopt |
△pykF/△pykA/△feaB |
△tyrR/aroGfbr/tyrAfbr |
– |
– |
SF |
Glucose |
0.06 |
[14] |
| Saccharomyces cerevisiae HLF‐Dα |
Bbxfpkopt/Pcaasopt/Atugt85a1opt |
EctyrAfbr |
△pdc1/△pha2/△trp3 |
EctyrAfbr |
– |
– |
FB (3 L) |
Glucose |
1.82 |
[24] |
| Saccharomyces cerevisiae CEN.PK2-1C |
Rru8gt33opt |
rki1/tkl1/aro2/aro10 |
△pdc1/△pha2 |
aro4fbr/aro7fbr/aro3fbr |
– |
– |
FB (5 L) |
Glucose |
9.90 |
[25] |
| Saccharomyces cerevisiae BY4742 |
Pcaas/Atugt85a1 |
EcaroL |
– |
aro4fbr/aro7fbr |
– |
– |
SF |
Glucose |
0.24 |
[37] |
| Saccharomyces cerevisiae BY4742 |
Pcaas/Atugt85a1 |
EcaroL |
– |
aro4fbr/aro7fbr |
– |
– |
FB (5 L) |
Glucose |
0.73 |
[37] |
| Escherichia coli MG1655 |
Atugt85a1 (eight copies)/Scaro10opt (five copies) |
– |
△feaB/△pykF/△pabA/△pabB/△typE/△pheA/△tyrB |
△tyrR |
– |
– |
SF |
Glucose |
2.42 |
[38] |
| Escherichia coli MG1655 |
Atugt85a1 (eight copies)/Scaro10opt (five copies) |
– |
△feaB/△pykF/△pabA/△pabB/△typE/△pheA/△tyrB |
△tyrR |
– |
– |
FB (5 L) |
Glucose |
9.34 |
[38] |
| Escherichia coli BL21(DE3) |
Rsugt72b14opt |
– |
– |
– |
– |
✔ |
SF |
Tyrosol |
0.01 |
[39] |
| Escherichia coli BL21(DE3) |
Blugt |
– |
– |
– |
– |
– |
SF |
Tyrosol |
1.04 |
[40] |
| Saccharomyces cerevisiae BY4743 |
Rr4hpaasopt/Rrt8gtopt |
– |
– |
aro4fbr/aro7fbr |
– |
– |
SF |
Glucose |
< 0.01 |
[41] |
| Escherichia coli-Escherichia coli |
Ppkdc4opt (E. coli-1)/Atugt85a1opt (E. coli-2) |
aroE (E. coli-1)/Pgm/galu (E. coli-2) |
△manz/△feaB/△mao/△ptsG/△pykA/△pheA (E. coli-1)/△xyla/△usha/△tyrA (E. coli-2) |
aroGfbr/tyrAfbr/△tyrR (E. coli-1) |
– |
✔ |
FB (5 L) |
Glucose |
6.03 |
[42] |
| Escherichia coli-Escherichia coli |
Ppkdc4opt (E. coli-1)/Atugt85a1opt (E. coli-2) |
aroE (E. coli-1)/Pgm/galu (E. coli-2) |
△manz/△feaB/△mao/△ptsG/△pykA/△pheA (E. coli-1)/△xyla/△usha/△tyrA (E. coli-2) |
aroGfbr/tyrAfbr/△tyrR (E. coli-1) |
– |
✔ |
FB (5 L) |
Glucose |
6.03 |
[42] |
| UGT: Uridine diphosphate glucosyltransferase; Atugt85a1: UGT from Arabidopsis thaliana; Rru8gt33: UGT from Rhodiola rosea; Rsugt73b6: UGT from R. sachalinensis; Rr4hpaas: 4HPAA synthase gene from R. rosea; Rsugt72b14: UGT from R. sachalinensis. The annotations for other gene abbreviations are consistent with Table 1. |
There are three main ways to obtain tyrosol: natural extraction, chemical synthesis, and microbial synthesis. Due to its excellent biological and pharmacological activity, the market demand for tyrosol is increasing. However, there are numerous challenges associated with natural extraction, such as a low extraction rate and the long growth cycle of plants[43]. On the other hand, chemical synthesis encounters complex reactions and an inhospitable environment[11]. Therefore, the construction of microbial cell factories for synthesizing tyrosol has become a potential industrialization method, which not only offers cost-effectiveness but also provides the advantages of environmentally friendly production, garnering significant attention in recent years.
Escherichia coli and Saccharomyces cerevisiae are common chassis cells for metabolic engineering. They have a clear genetic background, mature gene editing techniques, and effective high-density fermentation characteristics[44]. Especially for S. cerevisiae, it is generally recognized as a safe (GRAS) strain. It does not need to introduce an exogenous synthesis pathway but can synthesize tyrosol through its endogenous Ehrlich pathway[45].
Therefore, the use of E. coli and S. cerevisiae to synthesize tyrosol has aroused the interest of researchers.
As the tyrosol derivatives, including hydroxytyrosol and salidroside, have also been reported to possess various biological functions, such as anti-oxidation, anti-inflammation, and anti-virus[12,46], their synthetic pathway based on the biosynthetic pathway of tyrosol has also been successfully constructed and reasonably regulated in E. coli and S. cerevisiae.
In this review, the biosynthetic pathway of tyrosol is summarized. The key enzymes and nodes in the de novo synthesis pathway of tyrosol in E. coli and S. cerevisiae are analyzed. Furthermore, recent advancements in the synthesis of tyrosol by E. coli and S. cerevisiae are reviewed. Moreover, the key catalytic enzymes involved in the synthesis of hydroxytyrosol and salidroside are introduced, and the recent advancements in the metabolic engineering of these compounds are summarized. This review will provide a reference for constructing high-yield engineering strains producing tyrosol and its derivatives.
1 Application of metabolic engineering in the microbial synthesis of tyrosol and its derivatives The metabolic networks of important compounds in cell factories can be constructed and regulated by metabolic engineering, which has been used in various fields such as food, chemistry, medicine, and biology[47]. To achieve efficient microbial synthesis of tyrosol and its derivatives, metabolic engineering is mainly carried out from four aspects: (1) Mining the key rate-limiting enzymes in the pathway and identifying their related functions to construct the de novo synthesis pathway of tyrosol and its derivatives; (2) Regulating metabolic network through conventional metabolic engineering, including pathway engineering, promoter engineering, and cofactor engineering, and the metabolic flow of the target product was increased; (3) Clarifying potential rate-limiting targets of tyrosol and its derivatives through reverse metabolic engineering; (4) Developing dynamic regulatory gene circuits to rationally distribute carbon flux to achieve efficient synthesis of the target tyrosol-derived products in microorganisms. The following four aspects of the de novo synthesis of tyrosol and its derivatives by metabolic-engineered microorganisms are introduced and summarized in detail.
2 Biosynthesis of tyrosol
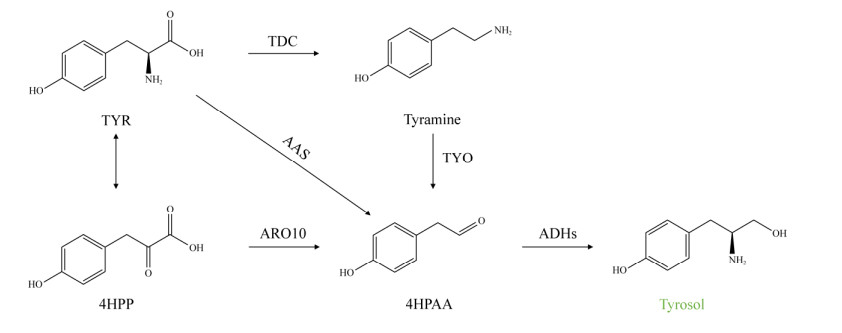
2.1 Biosynthetic pathway of tyrosol Three main biosynthetic pathways of tyrosol have been reported, all of which are derived from tyrosine: (1) Tyrosine can be converted to tyramine through the catalysis of tyrosine decarboxylase (TDC). Tyramine oxidized to 4-hydroxy-phenylacetaldehyde (4HPAA) by tyramine oxidase (TYO). Subsequently, 4HPAA is reduced by alcohol dehydrogenase (ADH) to form tyrosol[13,48]. (2) Tyrosol can be directly synthesized from 4HPAA under the catalysis of aromatic aldehyde synthase (AAS)[12]. (3) Tyrosol can be synthesized by the Ehrlich pathway: tyrosine is deaminated by aromatic amino acid aminotransferase (ARO8) and then forms 4HPAA by decarboxylated reaction through pyruvate decarboxylase (ARO10), and 4HPAA is reduced to tyrosol by alcohol dehydrogenase[49] (Figure 1).
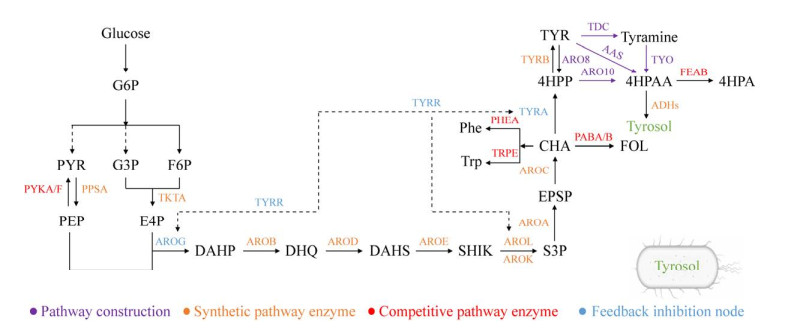
2.2 De novo production of tyrosol by metabolic engineering of E. coli E. coli, as a commonly used bacterial cell factory, has been widely utilized for microbial synthesis of various high-value compounds, such as tyrosol. Since tyrosol is not a natural product of E. coli, constructing a de novo synthesis pathway for tyrosol and implementing metabolic regulation are required to achieve a high yield of tyrosol in E. coli. The synthetic pathway and target enzymes of tyrosol in E. coli are shown in Figure 2, and the recent advances in the synthesis of tyrosol by metabolic engineering of E. coli are summarized in Table 1.
2.2.1 Construction of the de novo synthetic pathway of tyrosol in E. coli Since tyrosol is not an endogenous product in E. coli, multiple exogenous pathways have been attempted to be introduced into E. coli to achieve de novo synthesis of tyrosol. Currently, there are mainly three artificially constructed tyrosol synthesis pathways in E. coli (Figure 2). Satoh et al.[13] realized the transformation of tyrosine to 4HPAA by introducing TYO from Micrococcus luteus and TDC from Papaver somniferum into E. coli. 4HPAA was then converted to tyrosol by endogenous alcohol dehydrogenase through the TYO-TDC-ADHs pathway. Chung et al.[12] severally overexpressed aas from Arabidopsis thaliana, Petunia hybrid and Petroselinum crispum in E. coli to achieve the conversion of tyrosine to tyrosol; the results showed that AAS derived from Petroselinum crispum has the best performance, and the engineered strain produced 12.6 mg/L of tyrosol with the exogenous addition of 100 μmol/L of tyrosine. This indicates that introducing exogenous aas to E. coli achieved the direct transformation of tyrosine to 4HPAA, and the 4HPAA is reduced to tyrosol through the action of alcohol dehydrogenase (AAS-ADHS pathway)[11-12]. The third pathway involves the synthesis of based on the Ehrlich pathway ofS. cerevisiae. Bai et al.[14] overexpressed aro10 from S. cerevisiae in E. coli and successfully constructed a metabolic pathway from 4-hydroxyphenylpyruvate (4HPP) to 4HPAA, and then 4HPAA is converted to tyrosol by the endogenous ADHs (ARO10-ADHs pathway). The ARO10-ADHs pathway is highly efficient, and it is a common artificial metabolic pathway used by E. coli for the synthesis of tyrosol[14-21]. In addition, the codon preference of bacteria is significantly different from that of fungi. Yang et al.[20] found that the aro10 from the S. cerevisiae genome has many codons rarely used in the E. coli genome. After codon optimization based on E. coli preferences, the engineered strain showed enhanced expression levels of aro10 and increased production of tyrosol. Subsequently, the yield of tyrosol was significantly improved from 3.11 mmol/L to 10.92 mmol/L by increasing the copy number of aro10opt to five copies in the engineered E. coli[17]. This suggests that the ARO10-ADHS pathway can achieve de novo synthesis of tyrosol in E. coli, and ARO10 is an important regulatory target.
In addition to the three main synthetic pathways mentioned above, Lai et al.[22] conducted an enzyme cascade reaction in E. coli. The conversion of p-coumaric acid (p-CA) to tyrosol was achieved using ferulic acid decarboxylase from S. cerevisiae, styrene monooxygenase, and styrene oxide isomerase from Pseudomonas putida, and phenylacetaldehyde reductase from Solanum lycopersicum. The synthetic pathway from endogenous tyrosine to p-CA was established by introducing exogenous tyrosine ammonia-lyase from Flavobacterium johnsoniae. Finally, the synthesis of tyrosol from glucose, glycerol, and p-CA was achieved in E. coli, resulting in the establishment of a new synthesis pathway for tyrosol.
2.2.2 The key node in the tyrosol synthetic pathway To increase the yield of tyrosol in E. coli, overexpressing of the key enzymes involved in the synthetic pathway is a common strategy. In the biosynthetic pathway of tyrosol, phosphoenolpyruvate (PEP), an intermediate of glycolysis, and erythrose-4-phosphate (E4P), an intermediate of the pentose phosphate pathway, are two crucial precursors. PEP and E4P can be transformed into tyrosol through the shikimic acid pathway and the Ehrlich pathway. By enhancing the supply of PEP and E4P through overexpression of the genes encoding phosphoenolpyruvate synthase (ppsA) and transketolase (tktA), respectively, the production of tyrosol was increased in E. coli[14]. Various synthases in the shikimic acid pathway are also key enzymes in the tyrosol synthetic pathway. Bai et al.[14] modified multiple enzyme nodes in the shikimic acid pathway in E. coli, including overexpressing the gene aroE coding shikimate dehydrogenase, the gene aroD coding 3-dehydroquinate dehydratase, and the gene aroBopt coding 3-dehydroquinate synthase. The results showed that the control strain could produce 752.6 mg/L tyrosol, and the engineered strain overexpressing aroE increased the tyrosol yield to 824.9 mg/L, while the engineered strain overexpressing aroE, aroD, and aroBopt further increased the tyrosol yield to 926.9 mg/L[14]. This indicates that Shikimate dehydrogenase (AROE), 3-dehydroquinate dehydratase (AROD), and 3-dehydroquinate synthase (AROB) in the shikimic acid pathway are key rate-limiting enzymes for the synthesis of tyrosol. Similar results were also found in the study by Yang et al.[15]. Overexpression of 5-enolacetone shikimic acid-3-phosphate synthetase coded genes aroA and aroE had a positive impact on the tyrosol yield[15]. 4HPAA is a crucial precursor for the synthesis of tyrosol. Alcohol dehydrogenase plays a key role in the conversion pathway from 4HPAA to tyrosol, serving as a key pathway synthetase node. By enhancing the expression of endogenous alcohol dehydrogenase YAHK, the conversion rate of 4HPAA to tyrosol can be increased, thereby improving the yield of tyrosol[15,18].
2.2.3 The feedback inhibition node In E. coli, the activity of the inhibitor TYRR in the aromatic amino acid pathway can be regulated by binding to one or more aromatic amino acids[50]. In the presence of ATP and tyrosine, the TYRR protein can inhibit the transcription of genes encoding many key enzymes in the tyrosine synthesis pathway, such as the shikimic acid kinase gene aroL and the prephenate dehydrogenase gene tyrA, etc.[51-52]. Chung et al.[12] found that deleting the tyrR gene increased the production of tyrosine in E. coli. Since tyrosol is a derivative of tyrosine, the strategy of deleting tyrR has also been utilized in metabolic engineering of E. coli to produce tyrosol and attain the desired outcomes[11,12,14,17,19]. In addition, DAHP synthetase AROG and TYRA were inhibited by phenylalanine and tyrosine feedback, which was not conducive to the synthesis of aromatic amino acids. Therefore, inhibiting of the feedback pathway by introducing of aroGD146N and tyrAM53I/A354V into E. coli increased the metabolic flux of the shikimic acid pathway and leading to an enhancement in tyrosine synthesis[53]. In another study, the inhibitory feedback mutants aroGfbr and tyrAfbr were overexpressed in E. coli, which successfully increased the production of tyrosol from 603.9 mg/L to 752.6 mg/L[14].
2.2.4 The key node in the competitive pathway of tyrosol By eliminating or weakening the competitive pathway, we can avoid the waste of carbon metabolic flow and redirect more carbon flux towards tyrosol. As a key node, chorismate can generate tyrosine by the action of prephenate dehydrogenase (TYRA) and aromatic amino acid transaminase (TYRB) while also generate phenylalanine and tryptophan under the action of chorismate synthase (PHEA) and anthranilate synthase (TRPE), respectively. Therefore, in order to enhance the synthetic capability of tyrosol in E. coli, the pheA involved in the competitive pathway was knocked out, increasing the tyrosol yield from 0.14 mmol/L to 0.29 mmol/L[17]. 4HPAA is another key node in the synthetic pathway of tyrosol, which is utilized not only for synthesizing tyrosol under the action of alcohol dehydrogenase but also transformed into the byproduct ethyl 4-hydroxyphenylacetate (4HPA) with the action of phenylacetaldehyde dehydrogenase FEAB. Therefore, deletions of pheA and feaB are usually selected for the production of tyrosol[11-17,,20-22]. In addition to regulating the main competitive pathway, other competitive pathways were also engineered. PEP could be converted into pyruvate under the action of the pyruvate kinase isoenzyme PYKA/PYKF. To direct more PEP towards tyrosol, Bai et al.[14] and Xu et al.[17] deleted pykA/pykF to block the conversion of PEP to pyruvate, which improved the tyrosol synthesis ability in E. coli.
2.2.5 Dynamic regulation of metabolic flow Static regulation strategy is common and effective in metabolic engineering. However, the accumulation of certain target products and key intermediates can be toxic to cell growth, which is not conducive to achieving high yields of target products. Additionally, there is a conflict between product accumulation and cell growth in the competition of carbon sources. Static regulation strictly regulates metabolic flow, which is not conducive to the maximum utilization of carbon sources, while dynamic regulation strategies can compensate for the deficiency of static regulation[23]. Tyrosol has been found to have strong antibacterial activity, which can significantly impact the morphology and physiological state of yeast and bacteria[54-55]. In addition, Shen et al.[18] found that the engineered tyrosol-producing E. coli had a low-density problem, and studies showed that a high concentration of tyrosol had an inhibitory effect on the growth of E. coli. Therefore, Shen et al.[18] constructed a quorum sensing system to balance the synthetic pathway of tyrosol and the growth of bacteria, which increased the yield of tyrosol by 33.82% to 1.74 g/L. Moreover, the beneficial effect of the quorum sensing system dynamically regulating the synthesis of tyrosol in E. coli was particularly evident in high-density fermentation, and the tyrosol yield of the dynamically regulated engineering strain was 38.58% higher than that of the statically regulated engineering strain[18].
2.2.6 Optimization of fermentation environment In addition to metabolic engineering strategies, the regulation of the fermentation environment of engineered strains is also important to improve the yield of tyrosol. Xue et al.[16] investigated the influences of induced temperature, induced time, and the concentration of the inducer (isopropyl-β-D-thiogalactoside, IPTG) of recombinant protein. It was found that the induced temperature had an effect on the enzyme activity of the recombinant protein, while the duration of induction had a significant impact on the expression of the recombinant protein. The concentration of the inducer directly affects the activation of enzyme expression. Finally, the yield of tyrosol is improved by optimizing these factors. Similar optimization of fermentation environment factors for strains was also conducted by Chung et al.[12], Xue et al.[21] and Yang et al.[20], which enhanced the engineered strains' capacity to synthesize tyrosol.
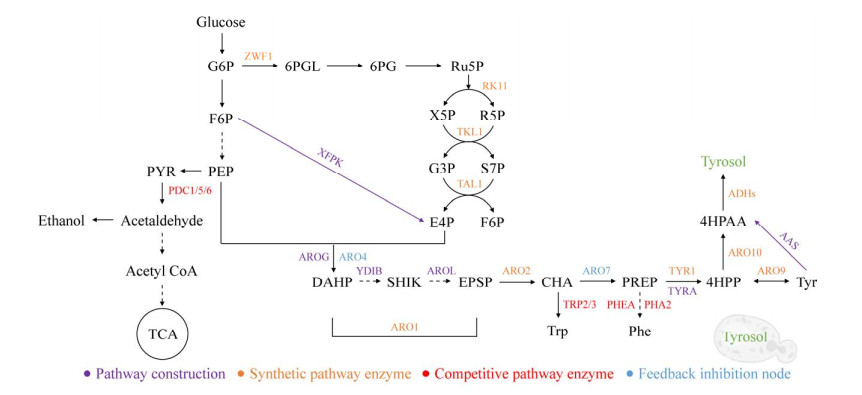
2.3 De novo production of tyrosol by metabolic engineering of S. cerevisiae S. cerevisiae was commonly chosen as a microbial synthesis chassis cell for producing tyrosol due to its safety and the endogenous Ehrlich pathway. The synthetic pathway and regulatory target enzymes of tyrosol in S. cerevisiae are shown in Figure 3. In this review, the recent advancements in the synthesis of tyrosol through metabolic engineering of S. cerevisiae are summarized in Table 1.
2.3.1 The pathway of de novo synthesis of tyrosol in S. cerevisiae In S. cerevisiae, glucose can be utilized to synthesize PEP and E4P through the glycolysis and pentose phosphate pathway, respectively. Subsequently, PEP and E4P can be converted into 4HPP via the shikimic acid pathway. Finally, 4HPP decarboxylates to produce 4HPAA under the catalysis of ARO10 in the Ehrlich pathway. 4HPP can be interconverted with tyrosine under the action of ARO8/9. To avoid the waste of carbon sources and maximize the utilization of carbon sources, Guo et al.[10] introduced AAS from Petroselinum crispum to construct a direct conversion pathway from tyrosine to 4HPAA. Therefore, tyrosol can be converted and synthesized by 4HPAA through the natural synthesis pathway of yeast (ARO10-ADHs pathway) and the artificial construction pathway (AAS-ADHs) simultaneously, leading to a significant increase in yield. According to previous reports, the flux of E4P in S. cerevisiae is significantly lower than that of PEP[56-58]. They are important precursors of tyrosol synthesis. The imbalance between E4P and PEP causes the low yield of tyrosol in S. cerevisiae. Therefore, Guo et al.[24] introduced the phosphotransferase XFPK from Bifidobacterium brevoidum into S. cerevisiae in a subsequent study on tyrosol, and they constructed the conversion pathway from fructose-6-phosphate (F6P) to E4P. Therefore, E4P can be synthesized by both the natural pentose phosphate pathway and the artificially constructed XFPK pathway. The flux of E4P has been increased, and the final yield of tyrosol is increased by 33.44%[24].
2.3.2 The key node in the tyrosol synthesis pathway The de novo synthetic pathway of tyrosol in S. cerevisiae mainly composed of the pentose phosphate pathway, shikimic acid pathway, and Ehrlich pathway. Reasonable modification of the pathway synthase in each pathway can increase the yield of tyrosol. The metabolic flux imbalance of E4P and PEP can be alleviated by the exogenous XFPK enzyme. In addition, Liu et al.[25] enhanced the anabolic flow of E4P by overexpressing genes encoding ribose-5-phosphate ketol-isomerase (RKI1) and transketolase (TKL1) in the pentose phosphate pathway to improve the yield of tyrosol. As expected, the overexpression of rki1 in the parent engineered yeast strains yielded 132 mg/L of tyrosol, which was 29.78% higher than that of the control strains. And overexpression of rki1 and tkl1 further increased the yield by 38.10%[25]. For the shikimic acid pathway, Liu et al.[25] separately overexpressed multiple pathway synthases: the five-functional enzyme ARO1, bifunctional chorismate synthase ARO2 and AROL from E. coli, which are responsible for the conversion of 3-deoxy-D-arabino-heptu-losonate-7-phosphate (DAHP) to 5-enolpyruvylshikimate-3-phosphate (EPSP), EPSP to chorismate (CHA), and shikimate (SHIK) to EPSP. However, only the overexpression of aro2 had a positive effect on improving tyrosol yield. In addition, tyrAM53I/A354V, a tyrosine feedback inhibitory mutant in E. coli, is responsible for the synthesis of CHA to 4HPP in the shikimic acid pathway. Studies have shown that overexpression of tyrAM53I/A354V in the synthesis of tyrosol in E. coli has a significant effect on the yield of tyrosol[24], so it is also introduced into yeast to improve the yield of tyrosol. The results showed that exogenous expression of tyrAM53I/A354V in S. cerevisiae also increased the yield of tyrosol[10,24]. In the Ehrlich pathway, ARO10 is responsible for converting 4HPP into 4HPAA, and then the 4HPAA is converted into tyrosol under the action of ADHs. Therefore, the aro10 is overexpressed by Liu et al.[25], which enhances the synthesis pathway from 4HPP to 4HPAA. This indicates that ARO10 is a key regulatory target of tyrosol in S. cerevisiae.
2.3.3 The feedback inhibition node In S. cerevisiae, 4HPP is a key node in the synthetic pathway of tyrosine and tyrosol, and the accumulation of 4HPP is significantly inhibited by the feedback of the 3-deoxy-7-phosphoheptulonate synthase ARO4 and the chorismate mutase ARO7. Therefore, the feedback-insensitive mutants aro4K229L and aro7G141S are overexpressed inS. cerevisiae to enhance the accumulation of 4HPP and thus increase the yield of tyrosine and its derivatives[27]. Liu et al.[25] found that overexpression of aro4K229L and aro7G141S in S. cerevisiae increased the yield of tyrosol by three times.
2.3.4 The key node in the competitive pathway of tyrosol S. cerevisiae is a fungus that can produce ethanol. In S. cerevisiae, PEP can be converted to ethanol step by step by enzymes, including pyruvate decarboxylase PDC1/5/6. As the metabolic flow of PEP is redirected, this pathway competes with the synthetic pathway of tyrosol. In the synthetic pathway of tyrosine/tyrosol, tryptophan and phenylalanine are also produced. Therefore, the synthetic pathways of tryptophan and phenylalanine are also competitive to the synthesis of tyrosine/tyrosol[59]. To increase the metabolic flow of tyrosine synthesis, PDC1 responsible for ethanol synthesis, anthranilate synthase (TRP3) responsible for tryptophan synthesis, and prephenate dehydratase (PHA2) responsible for phenylalanine synthesis were regulated. Deletion of pdc1, trp3 and pha2 resulted in 0.93 g/L of tyrosol by shake-flask fermentation and 8.48 g/L of tyrosol by feeding fermentation in a 3 L fermenter[24]. Similar results were also found in the study by Liu et al.[25]; the deletion of pdc1 in the engineered strain yielded 339 mg/L tyrosol, and on this basis, deletion of pha2 increased the yield of tyrosol to 522 mg/L.
2.3.5 Dynamic regulation of metabolic flow Given the adverse effects of tyrosol accumulation on yeast, our team developed a modified galactose (GAL) regulatory system based on GAL80 protein deletion to dynamically regulate tyrosol production in S. cerevisiae[27]. In S. cerevisiae, there is a natural galactose-induced GAL regulatory system and a glucose-inhibited Snf1 network, both of which can regulate GAL promoters. When galactose is added to the medium, the transcriptional inducible factor GAL3 binds to galactose and then binds with the inhibitory factor GAL80, releasing the transcriptional activator GAL4 to initiate a series of GAL promoters. To save costs, our team deleted GAL80 so that the GAL regulatory system was no longer regulated by galactose but instead by glucose concentration. Key genes such as aro4K229L, aro7G141S, Pcaasopt and Bbxfpkopt in the tyrosol synthesis pathway were regulated by GAL promoters to achieve the dynamic regulation of the synthetic pathway of tyrosol in S. cerevisiae. When the glucose concentration was high, the yeast strains grew. When the glucose concentration is low, critical pathway genes are expressed, and tyrosol production starts. The production of tyrosol was increased from 185 mg/L to 461 mg/L by implementing this dynamic control system[27].
2.4 Comprehensive regulation of metabolic pathway of tyrosol De novo synthesis of tyrosol can be achieved by mining key rate-limiting enzymes, introducing them into microbial cells, and identifying their functions. On this basis, multiple rate-limiting targets were coordinated to achieve metabolic flow, pulling in the direction of tyrosol synthesis. Yang et al.[20] constructed the de novo synthesis pathway of tyrosol in E. coli by introducing ARO10 from S. cerevisiae, and knocked the inhibitory factor TYRR, resulting in the titer of tyrosol reaching 3.57 mmol/L; then, the coded gene of the key rate-limiting enzyme ARO10 is over-expressed, and the titer can be increased from 3.57 mmol/L to 8.72 mmol/L; finally, the titer can reach 9.53 mmol/L by optimizing the fermentation conditions. Similarly, the comprehensive regulation of metabolic networks was also found in the study of Shen et al.[18] about the microbial production of tyrosol. They regulated the rate-limiting target of an artificially constructed tyrosol pathway by the fusion of ARO10F138L/D218G and alcohol dehydrogenase from different sources. The optimal combination is ARO10F138L/D218G and YahK, and the titer of tyrosol reaches 1.09 g/L[18]. Regulating the competitive rate-limiting target of the tyrosol metabolic network increased the titer of tyrosol by 21.15%[18]. Finally, the quorum sensing dynamic system was designed to regulate the metabolic network of tyrosol globally, obtaining 1.74 g/L of tyrosol in shake-flask fermentation and 4.22 g/L after fermentation in the fermenter[18]. Liu et al.[25] achieved de novo synthesis of tyrosol based on the Ehrich pathway of S. cerevisiae and coordinated multiple metabolic engineering to comprehensively regulate the metabolic network of tyrosol, including strengthening pathway engineering for endogenous pathway nodes, weakening pathway engineering for competitive pathways, and introduction of insensitive feedback inhibitors for multiple feedback inhibition nodes. They finally achieved the highest yield of tyrosol (9.9 g/L) in a 5 L fermenter[25].
3 Biosynthesis of tyrosol derivatives In the field of biochemistry, tyrosol can be used as an important intermediate to synthesize a variety of high-value compounds. The typical high-value tyrosol derivatives are hydroxytyrosol and salidroside. Owing to the antioxidant, anti-inflammatory, and anti-fatigue of hydroxytyrosol and salidroside[12,46], the market demand for these compounds is constantly increasing. Exploration of its green and efficient methods has gradually aroused the interest of researchers. In this review, the pathway enzymes involved in the synthesis of hydroxytyrosol and salidroside were summarized (Figure 4 and Figure 5), and the recent advantages in the synthesis of hydroxytyrosol and salidroside by metabolic engineering were reviewed (Table 2 and Table 3).
3.1 Biosynthesis of hydroxytyrosol
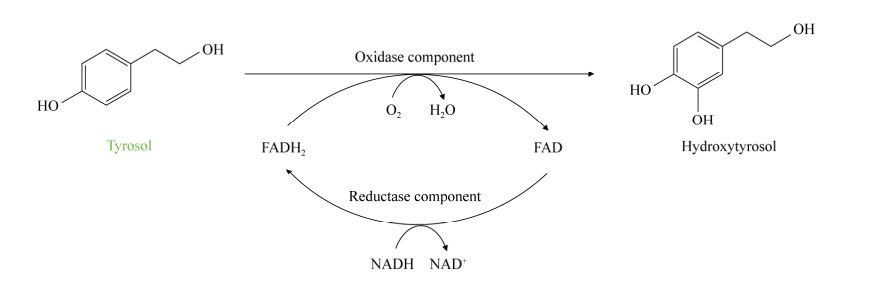
3.1.1 Tyrosol hydroxylase The conversion of tyrosol to hydroxytyrosol only requires one-step enzyme catalysis. The commonly used tyrosol hydroxylase is a two-component polyphenol oxidase, including the oxidase component and the reductase component. The mechanism of action is shown in Figure 4. During catalysis, the reducing enzyme component can transfer electrons from NADH to FAD to generate FADH2, and then FADH2 is captured by the oxidase component to achieve the hydroxylation of tyrosol in the presence of oxygen, and finally, FAD is released to achieve the cofactor cycle[27-28,60].
3.1.2 De novo production of hydroxytyrosol by metabolic engineering of microorganism As a derivative of tyrosol, the microbial synthesis pathway of hydroxytyrosol can be metabolically modified based on the biosynthesis pathway of tyrosol. Researchers constructed a product synthesis pathway from glucose to hydroxytyrosol in E. coli and then overexpressed key genes of the synthesis pathway as well as weakened the competitive pathway to regulate more metabolic flow to the direction of tyrosol synthesis. Finally, the transformation of tyrosol to hydroxytyrosol was enhanced through the overexpression of tyrosol hydroxylase[12,27-33]. Since the catalytic action of tyrosol hydroxylase HPABC requires the participation of cofactors, Wang et al.[28] introduced L-amino acid deaminase (LAAD) from Proteus mirus to promote the regeneration of FADH2 cofactors. Then, the engineered E. coli strains produced 3.12 g/L hydroxytyrosol from glucose and glycerol. The oxidase component of 4-hydroxyphenylacetate 3-monooxygenase (HPAB) is very important for the transformation of tyrosol, Qi et al.[33] successfully obtained a high conversion efficiency HPAB mutant HAPBS210T/A211L/Q212E/Y282H through directed evolution of HPAB. Since tyrosol is an endogenous product of S. cerevisiae, the microbial synthesis of hydroxytyrosol in S. cerevisiae has also been studied. Our study[23] found that the wild-type S. cerevisiae can only synthesize tyrosol, while no hydroxytyrosol product was detected in the fermentation of the wild-type S. cerevisiae. It indicates that the wild-type S. cerevisiae does not contain the tyrosol hydroxylase, which can not achieve de novo synthesis of hydroxytyrosol. Therefore, five hydroxylase sources were screened. The hydroxylase from Pseudomonas aeruginosa was found to display high catalytic activity in S. cerevisiae and used to construct the biosynthesis pathway of hydroxytyrosol in S. cerevisiae. Finally, the metabolic pathway was regulated by static and dynamic regulation strategy, achieving 308 mg/L of hydroxytyrosol[23]. Specifically, the implementation of static regulated engineering includes the following three modules: (1) Elimination of tyrosine feedback inhibition by overexpressing aro4K229L and aro7G141S; (2) Redistribution of metabolic flux of the tyrosine metabolic pathway by overexpressing Pcaasopt; (3) Balancing the supply between PEP and E4P by overexpressing Bbxfpkopt. The dynamic regulation strategy was carried out through a glucose-responsive dynamic regulation system, achieving a 6.88-fold improvement[23]. Liu et al.[34] constructed a de novo synthesis pathway for hydroxytyrosol in yeast strains by introducing HpaB from Pseudomonas aeruginosa (PaHpaB) and HpaC from E. coli (EcHpaC). To increase the titer of hydroxytyrosol, the competitive metabolic flow was weakened by down-regulating the expression of trp2, which is responsible for L-tryptophan biosynthesis, resulting in a 27.2% increase of hydroxytyrosol[34]. In addition, the cytosolic NADH supply was increased, which further increased the hydroxytyrosol titer by 36.9%[34]. Finally, a titer of 1.12 g/L of hydroxytyrosol was obtained in shaking flask fermentation, and 6.97 g/L of hydroxytyrosol was achieved after feeding fermentation[34]. It is the highest titer of hydroxytyrosol in S. cerevisiae reported at present. Wang et al.[28] introduced ARO10 and ADH into E. coli to construct a de novo synthesis pathway of hydroxytyrosol. The key rate-limiting nodes of the pathway, including endogenous hydroxylase nodes, competitive pathway nodes, and feedback inhibition nodes, were regulated by enhanced pathway engineering, weakened pathway engineering, and the introduction of mutation-insensitive enzymes, respectively. Finally, the current paper achieved the highest titer of hydroxytyrosol, 3.12 g/L by shake-flask fermentation and 9.87 g/L by fed-batch fermentation in a 5 L fermenter[28].
3.2 Biosynthesis of salidroside
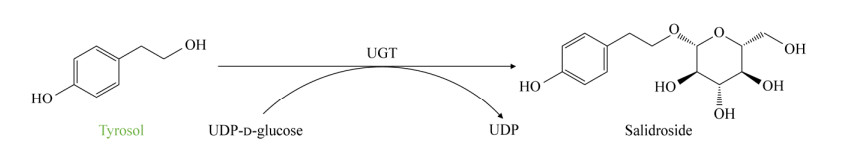
3.2.1 Uridine diphosphate glucosyltransferase By exploring the synthetic pathway of salidroside in plants, it was found that uridine diphosphate glucosyltransferase (UGT) is a key enzyme in the conversion of tyrosol to salidroside. In addition, uridine diphosphate glucose (UDPG) is required as a glycosylation donor in this glycosylation process[61-62].
3.2.2 De novo production of salidroside by metabolic engineering of microorganism The key to the synthesis of salidroside by microbial platforms is pathway reconstruction. As an aglycon of salidroside, tyrosol can be synthesized with UDPG under the action of the UGT enzyme. Bai et al.[14] achieved de novo synthesis of salidroside in E. coli by introducing the plant-derived UGT enzyme-coded gene Rsugt73b6opt. Other plant-derived UGT enzyme ATUGT85A1 has also been shown to achieve de novo synthesis of salidroside in engineered tyrosol-producing strains[12,63]. Liu et al.[38] enhanced the stability of the strains by integrating the codon-optimized atugt85a1opt into the genome of engineered tyrosol-producing E. coli to obtain 120 mg/L of salidroside. By adjusting the copy number of atugt85a1opt to eight, the yield of salidroside in the shaker was as high as 2.42 g/L, and the yield was as high as 9.34 g/L after feeding fermentation[38]. ATUGT85A1 was also introduced into S. cerevisiae to successfully construct the pathway of conversion from tyrosol to salidroside, and realized de novo synthesis of salidroside inS. cerevisiae. Since tyrosol is an important intermediate, key regulatory targets in the synthesis pathway of tyrosol in S. cerevisiae are also applicable to the regulation of the metabolic synthesis pathway of salidroside. To achieve the synthesis of salidroside, Liu et al.[25] introduced the UGT enzymes from three sources into the engineered yeast strains respectively for the production of tyrosol, including RsUGT72B14 from Rhodiola sachalinensis, RrU8GT33 from Rhodiola rosea and BsYjiC from Bacillus subtilis. The strain carrying RrU8GT33 had the highest yield of salidroside (1 575.45 mg/L) in shake flask culture[25]. After fed-batch fermentation, 9.9 g/L of salidroside was obtained in S. cerevisiae, which is the highest yield reported by now[25].
3.3 Biosynthesis of other derivatives In addition to the typical derivatives, such as hydroxytyrosol and salidroside, tyrosol can also be used as a substrate to synthesize other high-value compounds in microbial cell factories, including the acetylation product of tyrosol, icariside D2, verbascoside and osmanthuside B. According to reports, acetylation of phenolic compounds can enhance the lipophilicity and cell permeability of phenolic compounds and thus improve their bioavailability. Therefore, Guo et al.[64] achieved the acetylation of tyrosol by overexpressing the alcohol acetyltransferase coding gene atf1 in E. coli. To obtain more products, they first improved the de novo synthesis titer of the precursor tyrosol by screening aldehyde reductase, and then overexpressed the critical path enzyme ATF1 in an engineered strain. The final de novo synthesis titer of the tyrosol acetylation product was 508 mg/L[64]. On this basis, the acetylation product of hydroxytyrosol was obtained by overexpression of hpaBC, and the titer was 226 mg/L[64]. In addition, icariside D2, as a natural glycoside of plant origin, is widely used in the treatment of cardiovascular disease, hypertension and inflammation. Icariside D2 can be specifically synthesized in E. coli by the glycosylation of tyrosol via the exogenous enzyme RrUGT3 derived from Rhodiola rosea. Liu et al.[65] achieved a 3.80 g/L titer of icariside D2 using glucose as the sole carbon source in E. coli by optimizing the fed-batch fermentation conditions. To balance the pathway metabolic flow, Liu et al.[65] assigned the biosynthetic pathway of icariside D2 to two E.coli and achieved de novo synthesis of icariside D2. Under the condition of a dual carbon source (glucose-xylose), the de novo synthesis titer of icariside D2 was 2.92 g/L[65].
Verbascoside is a well-known phenylethanoid glycoside with antioxidant, anti-inflammatory and neuroprotective activities. Yang et al.[66] clarified the biosynthetic pathway of verbascoside and its downstream product osmanthuside B by mining key rate-limiting enzymes, as well as then conducting transcriptomic and in vitro enzyme analysis of the extracted enzymes. The two key rate-limiting enzymes is a BAHD acyltransferase (hydroxycinnamoyl-CoA: salidroside hydroxycinnamoyltransferase) and a CYP98 hydroxylase (osmanthuside B 3, 30-hydroxylase), which were introduced into E. coli and thus microbial synthesis of osmanthuside B was realized based on the biosynthesis pathway of tyrosol. However, the current yield is relatively low, and the de novo synthesis yield is only 22 mg/L after metabolic regulation[66].
4 Conclusion and prospects As a water-soluble phenolic compound, tyrosol has a series of biological and pharmacological activities, such as anti-oxidation and neuroprotection. It can be used as an intermediate to synthesize high-value derivatives such as hydroxytyrosol and salidroside. Therefore, the synthesis of tyrosol and its derivatives by constructing microbial cell factories through metabolic engineering has become a green, efficient and low-cost way. In this review, the biosynthetic pathway of tyrosol and its recent advantages in common chassis cells including E. coli and S. cerevisiae are summarized. The key pathway enzymes and regulatory nodes for de novo synthesis of tyrosol are introduced. Moreover, the key catalytic enzymes of hydroxytyrosol and salidroside are introduced in combination with the previous research work of our laboratory. The recent advantages in the metabolic engineering of hydroxytyrosol and salidroside were also reviewed.
Up to now, tyrosol and its main derivatives, including hydroxytyrosol and salidroside, have achieved grams per upgrade. However, they are still far from industrialization. To increase the production of tyrosol and its derivatives, the future can be carried out from three perspectives: (1) Improve the growth status of strains, studies have shown that the accumulation of tyrosol and its derivatives will have adverse effects on cells, and further studies can be conducted to improve the robustness and stress resistance of strains; (2) Realize dynamic regulation of metabolic network and rational utilization of carbon sources. The response elements of the gene circuit can be mined to build a dynamic regulatory system to maximize the utilization of carbon atoms. (3) Explore potential targets to achieve yield improvement. Omics-related tools can be used to explore potential targets of the metabolic network of target products, and develop reverse metabolic engineering to remove rate-limiting targets to increase the titer. In addition, the strains can also be macro-regulated. This can be done through whole-genome-directed evolution. The metabolic system can be regulated to improve production by mining response elements.
 2024, Vol. 40
2024, Vol. 40